The role of long noncoding RNAs in human T CD3+ cells
Manuela M Almo1, Isabel G Sousa2, Andréa Q Maranhão2,3, Marcelo M Brigido2,3*
1Molecular Pathology Graduation Program, Medicine Faculty, University of Brasilia, Brasilia, Brazil
2Department of Cell Biology, Institute of Biological Sciences, University of Brasilia, Brasilia, Brazil
3Institute for Immunology Investigation, a National Institute of Science and Technology
Abstract
Long noncoding RNAs (lncRNAs) are regulatory RNA molecules that are involved in various biological processes. In the immune system, the lncRNAs play important roles in development, differentiation, survival, cell fate determination, proliferation and activation of immune cells. Lymphocytes are the main players of the adaptive immunity and CD3+ T cells acts as a master regulator for the immune responses. These cells following activation by antigens and co-stimulatory signals are differentiated into various effector T cell subsets, including CD4 and CD8 T cells. These heterogeneous populations can be distinguished based on molecular surface markers and subsets of these markers can be used to denote various stages of T lymphocyte differentiation, notwithstanding the CD3+ T cells phenotypes are markedly influenced by lncRNAs. In the present review, we summarize recent research on the role of long noncoding RNAs in subtypes of CD4+ and CD8+ human T cells.
Noncoding RNAs (ncRNAs) are regulatory RNA molecules that do not code for proteins even though they affect and regulate a range of biologic processes. The ncRNAs can control several genes and are classified according to their size. They may be small, such as microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs), or long, such as long non-coding RNAs (lncRNAs) and long intergenic ncRNA (lincRNA)1. Long noncoding RNAs (lncRNAs) are defined as greater than >200 nucleotides in length, and the most of those currently described are transcribed by RNA polymerase II, therefore they are polyadenylated, and harbor the N-terminal added of a methylated guanosine nucleotide (5 'cap)2.
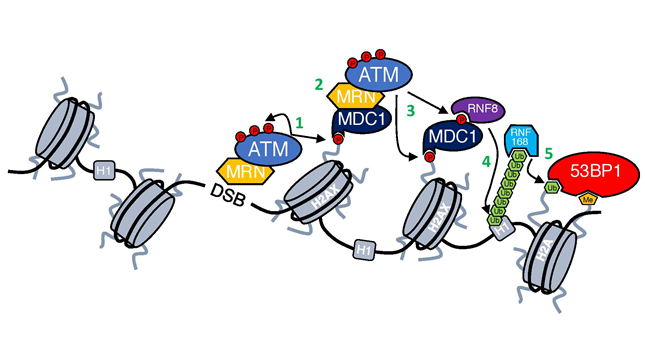
Figure 1: DNA damage response cascade.
Upon recruitment to DSBs, ATM dimer undergoes a complicated process of activation. First, it is acetylated by lysine acetyltransferase 5 (KAT5/TIP60) at Lysine 301615, which is followed by autophosphorylation, in-trans on Serine 1981, leading to its monomerization and conferring access of ATM substrates to the kinase domain16,17(Figure 2A).
The positioning of the lncRNAs genes in the genome is the parameter used to further classify these molecules, which can be divided into categories, such as: sense - lncRNAs that overlap one or more exons of a given transcript on the same strand; antisense - lncRNAs that overlap one or more exons of a transcript on the opposite strand; divergent - lncRNAs that initiate transcription on the opposite strand of a gene; bidirectional - when their expression and the expression of a neighboring coding transcript is initiated in close proximity; intronic - when they lay in an intron; intergenic - if they lie within a sequence between two genes3,4,5 .
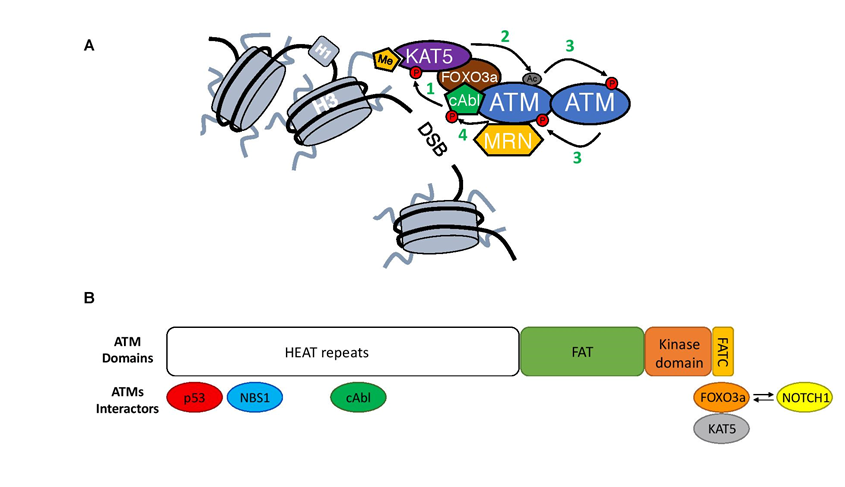
Figure 2: Close-up on ATM activation.
The progress in the studies of transcriptomes allowed the discovery and characterization of several ncRNAs and deepened knowledge about the involvement of these molecules in various human biological pathways. These studies also showed that ncRNAs regulate the gene expression at transcriptional, posttranscriptional and epigenetic levels3.
It is currently admitted that the differentiation of several lineages of the immune system are under regulation of lncRNAs6, such as monocytes, macrophages, dendritic cells, neutrophils, B and T lymphocytes7. Lymphocytes are the main regulators cells of the adaptive immunity and CD3+ T cells play a vast role in immune responses. These cells following activation by antigens and co-stimulatory signals are differentiated into various effector T cell subsets, including CD4 and CD8 T cells.
T cells play a central role in the regulation of the immune system and part of this function is performed by lncRNAs. In the thymus, each stage of differentiation of T cells is characterized by a unique pattern of expression of specific lncRNAs8. The identification of lncRNAs and their target genes that regulate T lymphocytes is important for the development of new therapies for autoimmune diseases, allergies, organ transplantation and cancer4. The purpose of this review is to describe the diversity of mechanisms used by lncRNAs to regulate T cell function and differentiation (Table 1).
NRON
The ncRNA repressor of NFAT is a lncRNA that inactivates the transcription factor NFAT under normal organism conditions. NFAT is a Ca2+ dependent transcription factor and controls the IL-2 production in activated T cells. In non-activated cells, NFAT is phosphorylated and inactive6. In response to increased intracellular Ca2+, the calcineurin enzyme dephosphorylates NFAT, allowing its translocation to the nucleus, where it will activate transcription of target genes, such as IL-28. NRON is part of a ribonucleoprotein complex that retains the NFAT factor in the cytoplasm in its phosphorylated form, preventing it from nuclear translocation. NRON is essential to maintain the structure of this protein complex, since in the absence of NRON the complex is not formed, and NFAT is dephosphorylated and routed to the nucleus where it performs its role as a transcription factor6,8,9.
NEST
NeST, also known as IFNG-AS1 or Tmevpg1, is a lncRNA expressed in CD4+, CD8+ T and NK cells10. Expressed in the Th1 cell, this lncRNA is an antisense transcript in the locus encoding IFNγ, acting in cis, as an enhancer, to stimulate the expression of IFNγ11. Once NeST is involved in the regulation of Th1 differentiation, this lncRNA depends on the expression of T-bet and STAT4 transcription factors12.
NeST binds to WDR5 subunit of the histone H3K4 methyltransferase complex altering by epigenetic mechanisms, methylation of the locus of the IFNγ gene, regulating its expression13.
Linc-MAF-4
Another lncRNA involved in T cell differentiation in Th1 lineage is MAF-4 (linc-MAF-4) long intergenic noncoding RNA. Linc-MAF-4 represses the expression of the MAF gene that in turn, encodes a transcription factor that promotes the expression of genes encoding specific cytokines of the Th2 line of CD4+ T lymphocytes14.
Linc-MAF-4 disrupts the differentiation of T cells into Th2 and promotes Th1 lineage differentiation6. Linc-MAF-4 and MAF genes lays close to each other in the human chromosome 16, which facilitates the action of the linc-MAF-4 in cis to regulate gene expression6. Linc-MAF-4 associates with chromatin modifiers, such as LSD1 and EZH2, to facilitate the trimethylation of histone H3K27 markers in the MAF gene promoter, by forming a loop in the chromatin, and thereby silencing its expression in Th1 cells8.
Th2LCRR
The lncRNA Th2LCRR is co-expressed with the cytokines that characterize Th2 polarization: IL-4, IL-5, IL-13 and is transcribed as an antisense of the RAD50 gene15. This lncRNA binds to the WDR5 component of the histone H3K4 methyltransferase complex to maintain the epigenetics markers in the interleukins IL-4, IL-13, IL-15 promoters, and thereby regulate T cell polarization8.
lincR-Ccr2-5'
The lincR-Ccr2-5 'AS is transcribed in the antisense direction of the Ccr2 gene and controls the expression of genes related to the response of the Th2 subtype11. The knockout of this lincRNA negatively alters the expression of chemokine receptor genes such as Ccr1, 2, 3 and 5, which are located at the same genomic loci as the non-coding intergenic Ccr2-5'AS16.
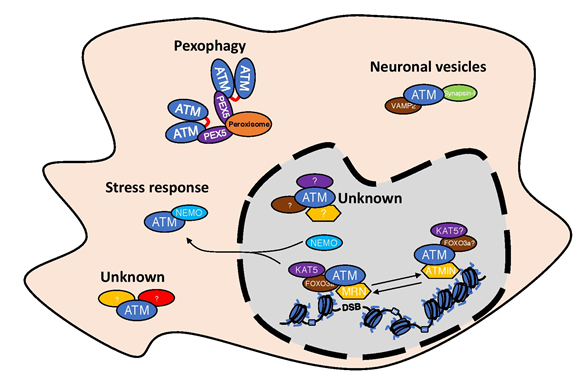
Figure 3: Existence of different ATM complexes in the cells.
GATA3-AS1
Another lncRNA characteristic of Th2 response, is the divergent lncRNA GATA3-AS117. Transcribed in both human and mouse genome, GATA3-AS1 is involved in allergic or asthmatic responses in humans, having its expression induced by allergens15. The involvement of GATA3-AS1 in certain forms of breast cancer and acute lymphoblastic leukemia in children has also been studied8.
RMRP
T cells that differentiate into the Th17 subtype are involved in the pathogenesis of autoimmune diseases and play an important role in the defense against extracellular pathogens18. The transcription factor RORγt is responsible for inducing this differentiation pathway, along with the cytokines IL-6 and TGF-β. RORγt can be activated by the DEAD box RNA helicase DDX5, an important catalyst for unfolding of RNAs, and this association, regulates the transcription of genes involved in the Th17 response6.
The interaction of RORγt factor with DDX5 requires the RMRP lncRNA (RNA component of the Mitochondrial RNase complex) within the nucleus. This DDX5-RMRP complex is essential for regulating the effector function of RORγt in several Th17 specific genes. LncRNA acts in trans in the genes dependent on this transcription factor, with the aid of the DDX5 helicase19.
The discovery of this RMRP-DDX5 complex in the regulation of RORγt showed two important factors: the complexity in Th17 cells and the possibility of other immune cells using the association of ncRNAs with RNA helicases to control gene expression20.
Flicr
Recent studies have shown the presence of lncRNAs in regulatory T cells (Tregs). Flicr (FOXP3 long intergenic noncoding RNA) is a lncRNA that negatively regulates the expression of FOXP3 transcription factor in regulatory T cells of humans and mouse21,22. Zemmour et al., suggests that the regulatory mechanism of lncRNA repression is related to changes in local accessibility of the FOXP3 gene in its conserved noncoding sequence 3 (CNS3) by controlling regulatory T cells activity, Flicr can trigger autoimmune diseases such as diabetes21. Since it is a newly discovered lncRNA, there is still scarce information concerning its functions and mechanisms of action.
HULC
Another lncRNA associated with regulatory T cells, is the lncRNA HULC (highly up-regulated in liver cancer). First described by Panzitt et al., in hepatocellular carcinoma (HCC), this cytoplasmic lncRNA plays a role in the posttranscriptional modulation of gene expression in HCC cells23. HULC was also studied in hepatitis B virus (HBV) related liver cirrhosis, an infection that can progress to the liver fibrosis stage, with an increase of CD4+ T cells, as described by ZHAO et al. These authors also suggested that the lncRNA HULC could trigger the regulatory T cells differentiation in HBV related liver cirrhosis. Also, HULC can down regulate the levels of the p18 protein that impairs the differentiation of regulatory T cells24.
lnc-EGFR
EGFR (epidermal growth factor receptor), a component of the receptor tyrosine kinase family, plays a role in the development of cancer, acting in the tumor microenvironment regulating the immune system cells25. An EGF Receptor binding RNA, Lnc-EGFR, seems to block its ubiquitination, triggering a cascade of activation, differentiation or suppression of certain cells25. Among T lymphocytes, the lnc-EGFR was shown to be upregulated in regulatory T cells in HCC patients. Also, it can stimulate the differentiation of regulatory T cells, as well as to suppress the activity of cytotoxic T cells, stimulating tumor growth. JIANG et al., suggested that the interaction between lnc-EGFR and regulatory T cells is a common pathway during the tumorigenesis process25.
Linc-POU3F3
Linc-POU3F3 is a lncRNA found in T regulatory cells of gastric cancer patients. When it is upregulated, this lncRNA reorganizes T regulatory cells distribution, allowing gastric cancer cells proliferation. This mechanism occurs through recruitment of TGF-β and the activation of its signaling pathway. These characteristics may pave the way for further exploration of lncRNAs function in immunotherapy of gastric cancer26.
NTT
NTT (non-coding transcript in T CD4+ cells) was one of the first lncRNAs identified in immune cells, being expressed in activated human T CD4+ cells11. Although its function has not yet been defined, it is known that there may be a link between NTT and the IFN-γ Receptor gene, since they share the same genomic locus and have similar expression profile during the activation of T lymphocytes11,27.
LincRNA-p21
LincRNA-p21 was first described to be involved with the p53 protein in response to DNA damage, however, it is also known that these lincRNA has its reduced levels in inflammatory diseases such as rheumatoid arthritis (RA) and coronary artery disease, which depend on the T cells8. Spurlock et al. demonstrated the role of lincRNA-p21 as a negative regulator of NF-κB activity in RA patients, where they also suggested that the expression of these lincRNA in RA can contribute to disease-dependent inflammation28.
CD8+ associated lncRNA
In a study using custom microarray probes, Pang et al. unveiled the expression profile of various lncRNAs in mammalian T CD8+ lymphocytes, that have not yet been well characterized in the context of CD8 T cell immunity. Most of these molecules are lymphoid-specific and may undergo changes depending on events that occur within lymphocytes, such as differentiation or activation29.
CD244 lncRNA
Some lncRNAs are not exclusive to T CD4+ cells, but their functions influence the development and activation of these cells, such as CD244 lncRNA. The CD244 costimulatory receptor regulates some functions of natural killer cells, is present in CD8+ T cells, and its persistent signaling is associated with some human viral infections such as Hepatitis B and C30. In tuberculosis, this receptor is involved in inhibiting the expression of cytokines such as IFN-γ and TNF-α, by inducing the expression of the lncRNA-CD244, that regulates the repression of the IFNG and TFN genes by epigenetic mechanisms. This lncRNA may represent an important target for therapeutic intervention in tuberculosis8,30.
Studies on the functions and mechanisms of action of lncRNAs are becoming trendy. Involved in several pathologies and related to various cellular subtypes, as was shown here, these molecules are being proposed as targets of therapies and diagnoses for diseases such as infections, autoimmune diseases, cancer8, ashtma and severe allergic disease2. However, questions such as how the lncRNAs interfere with the pathogen-host relationship, or how they act in various cellular processes still need to be established9.
Conclusion
In this review we updated the involvement of several lncRNAs in CD3+ T cells, where they are involved in development, differentiation, activation and effector functions. As a relatively new subject, several lncRNAs, mechanisms of action and functions have not yet been elucidated. However, advances in bioinformatics studies and new experimental analysis technologies may facilitate the discovery of new lncRNAs and a deeper understanding of those that are already known. For the immune system, lncRNAs may be important therapeutic targets for the treatment of various diseases, such as autoimmunity, infectious disease and even cancer.
Conflit of interest
The authors declare no conflict of interest.
Funding information
This work was perfomed under support of Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Apoio a Pesquisa do DF (FAP-DF) financial support.
References
- GUAN D Zhang W, Zhang W, et al. Switching cell fate, ncRNAs coming to play. Cell Death and Disease. 2013; v. 4(1: p. e464.
- AUNE TM, SPURLOCK CF. Long non-coding RNAs in innate and adaptive immunity. Virus Res. 2016; v. 212: p. 146–160.
- CHEN M, CHEN J, ZHANG D. Exploring the secrets of long noncoding RNAs. Int J Mol Sci. 2015; v. 16(3): p. 5467–5496.
- PAGANI, M. Rossetti G, Panzeri I et al. Role of microRNAs and long-non-coding RNAs in CD4+ T-cell differentiation. Immunol. Rev., 2013, v. 253(1), p. 82–96.
- DERRIEN T. The GENCODE v7 catalogue of human long non-coding RNAs?: Analysis of their structure , evolution and expression. Gen Res. 2012; v. 22: p. 1775–1789.
- ATIANAND MK, CAFFREY DR, FITZGERALD KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol. 2017; .v. 35: p. 177–198.
- BHAT SA. Long non-coding RNAs: Mechanism of action and functional utility. Non-coding RNA Res. 2016.
- AUNE TM, CROOKE PS, SPURLOCK CF. Long noncoding RNAs in T lymphocytes. J Leukoc Biol. 2015; v. 99(1): p. 31–44.
- GENG H, TAN XD. Functional diversity of long non-coding RNAs in immune regulation. Genes & Diseases. 2016; v. 3(1): p. 72–81.
- VIGNEAU S, Rohrlich PS, Brahic M, et al. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003; v. 77(10): p. 5632–8.
- ATIANAND MK, FITZGERALD KA. Long non-coding RNAs and control of gene expression in the immune system. Complement. 2015; v. 20(11): p. 623–631.
- COLLIER SP, Collins PL, Williams CL, et al. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. J Immunol. 2012; v. 189(5): p. 2084–2088.
- GOMEZ JA, Wapinski OL, Yang YW, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013; v. 152(4): p. 743–754.
- RANZANI V, Rossetti G, Panzeri I, et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015; v. 16(3: p. 318–25.
- SPURLOCK CF, Tossberg JT, Guo Y, et al. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. 2015; v. 6: p. 6932.
- HEWARD JA, LINDSAY MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014; v. 35(9): p. 408–419.
- ZHANG H, Nestor CE, Zhao S, et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J. Aller. Clinic. Immunol., 2013; v. 132(4): p. 1005–1008.
- DONG C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Ver Immunol., 2008; v. 8(5): p. 337–348.
- HUANG W, Thomas B, Flynn RA, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015; v. 528(7583): p. 517–522.
- ELLING R, CHAN J, FITZGERALD KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Europ J Immunol. 2016; v. 46(3): p. 504–512.
- ZEMMOUR D, Pratama A, Loughhead SM, et al. Flicr , a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Nat Acad Sci. 2017; p. 201700946.
- MOWEL WK, Kotzin JJ, McCright SJ, et al. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol. 2017; v. xx: p. 1–15.
- PANZITT K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a Novel Gene With Striking Up-Regulation in Hepatocellular Carcinoma, as Noncoding RNA. Gastroenterology. 2007, v. 132(1); p. 330–342.
- ZHAO J, Fan Y, Wang K, et al. LncRNA HULC affects the differentiation of Treg in HBV-related liver cirrhosis. Internat Immunopharma. 2015; v. 28(2): p. 901–905.
- JIANG R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017; v. 8: p. 1–15.
- XIONG G, Yang L, Chen Y, et al. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. Amer J Transl Res. 2015; v. 7(11): p. 2262–2269.
- LIU AY, Torchia BS, Migeon BR, et al. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics. 1997; v. 39(39): p. 171–184.
- SPURLOCK CF, Tossberg JT, Matlock BK, et al. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis and Rheum. 2014; v. 66(11): p. 2947–2957.
- PANG KC, Dinger ME, Mercer TR, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009; v. 182(12): p. 7738–48.
- WANG Y, Zhong H, Xie X, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8 + T-cell immune responses in tuberculosis infection. Proc Nat Acad Sci. 2015; v. 112(29): p. E3883–E3892.
