Prothymosin α, a protein implicated in the proliferation and survival of lymphocytes
Manuel Freire1*, Pablo Barbeito1, Concepción S. Sarandeses1, Cristina Díaz-Jullien1, Juan Muras1, Guillermo Covelo1, David Moreira1 and Carmen Freire-Cobo1
1The Department of Biochemistry and Molecular Biology, CIBUS, Faculty of Biology, University of Santiago de Compostela, 15782 Santiago de Compostela, Spain
Abstract
Prothymosin α (ProTα) is a 109-11 amino acid protein widely distributed in mammalian tissues and particularly abundant in lymphoid cells. Genomic and proteomic studies led to consider ProTα as a multifunctional protein implicated in nuclear and cytoplasmic functions. The nuclear function of ProTα is related to chromatin activity through its interaction with core histones and proteins involved in chromatin remodelling, whereas, processes related to the phosphorylation, the proteolytic processing to generate Thymosin α1, and the role as anti-apoptotic factor of ProTα, are linked to its cytoplasmic location. Affinity chromatography and co-immunoprecipitation experiments have demonstrated novel interactions of ProTα with acidic proteins such as SET, ANP32A, and ANP32B in the cytoplasm of proliferating lymphocytes. The stabilization of these interactions by chemical cross-linking with formaldehyde shows that they are formed through associations in six acidic complexes which correspond to selective interactions of SET and ANP32 proteins with ProTα. These ProTα-complexes also include cytoplasmic proteins implicated in membrane remodelling and in mitochondrial activity. In conclusion, these novel protein interactions of ProTα observed in proliferation activity and apoptosis studies, suggest that they might be related to mechanisms involved in the proliferation activity and the apoptotic control of lymphocytes.
Introduction
In 1966 White and Goldstein´s group intended to identify protein factors which could be implicated in the biological function of the thymus and thus isolated a mixture of polypeptides named thymosin1. The best known thymosin preparation is a calf thymus extract named Thymosin Fraction 5 (TF5). The TF5 contains about 30 different heat-stable polypeptides showing immune regulatory properties in several assay systems2 in vitro and in vivo. The first purified, sequenced and most characterized component of TF5 is the Thymosin α1 (Tα1), a polypeptide comprised of 28 amino acids. Tα1 is 10-1000 times more active than TF5 in different immune assays3. Thymosin α11 (Tα11), another component of TF5 structurally related to Tα1, was later isolated and sequenced4. The structure of Tα11 corresponds to the sequence of Tα1 plus seven additional amino acid residues at its C-terminus. Tα11 is about four times less abundant than Tα1 in TF5, however, it shows similar activity to Tα1 in some in vivo assays5.
The strategy to characterize a precursor of the α-thymosins in the thymic cells from translation products of calf thymus mRNAs led our group to isolate a protein which included Tα1 in its sequence6,7. This protein, isolated from rat thymus was later sequenced and named Prothymosin α (ProTα)8. The sequence of ProTα contains a Tα1 structure located at its first 28 amino acid residues and a Tα11 structure located at its 35 amino acid residues. The primary structure of ProTα obtained from other mammalian tissues demonstrates that ProTα is an acidic protein with a highly conserved sequence containing between 109 and 111 amino acids. Figure 1a shows the sequence of human ProTα9, which includes a central acidic region (residues 40-82) with glutamic and aspartic residues and other structural characteristics.
Later studies on ProTα gene expression end the theory of an exclusive role of the α-thymosins in the thymus immune function, as deduced from its generalized presence in different mammalian tissues10,11. The functional ProTα gene (PTMA) is located on chromosome 2 in humans12. A significative feature of ProTα is its high level of gene expression in proliferating cells13,14, on the range of core histones, especially in lymphoid tissues11. In Figure 1b are summarized the concentrations of ProTα in different rat tissues.
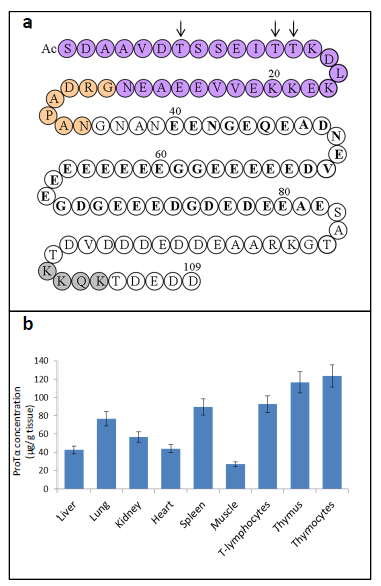
Figure 1. a) Human Prothymosin α amino acid sequence (1-109)9. AcS indicates acetylated N-terminal serine derived from the generalized co-translational processing which undergoes ProTα16. Tα1 sequence is indicated in purple circles. Tα11 sequence comprises the Tα1 sequence plus an additional 7 residues at its C-terminus (indicated in orange circles). Other structural characteristics of the ProTα sequence (referred in the text) are also indicated: arrows mark the phosphorylation sites of ProTα in normal (Thr7 and Thr12 or 13) and carcinogenic (only Thr7) lymphocytes; acidic region is indicated in bold type and nuclear localization signal is shown in grey circles. b) Concentrations of ProTα determined in extracts of different rat tissues11.
The biological function of ProTα
Further research on the intracellular evolution of ProTα provides important clues on its possible biological role. Characteristics of its gene structure and expression show the absence of a signal peptide15. Similarly, co-translational acetylation of its N-terminal serine residue16 indicates that ProTα should have an intracellular site of action or at least that it can be secreted by a non-conventional mechanism. Studies on the subcellular distribution of ProTα demonstrate a preferential nuclear location in proliferating cells17,18 which is in agreement with the presence of a nuclear localization signal at the C-terminus of its sequence19. However, the prominent cytoplasmic presence of ProTα in cell lysates demonstrates a special ability of this protein to leak from the nucleus during the cell extraction process15,20.
The characterization of the components present in a nuclear-protein complex isolated by affinity to ProTα20,21 has served towards the elucidation of the putative mechanisms by which ProTα might be involved in the nuclear activity. This nuclear-protein complex is comprised of core histones, acetyltransferases of histone H3 and H4, the methyltransferase of histone H3, as well as transcription factors and other proteins implicated in chromatin remodelling22. Its presence in this complex supports a nuclear role of ProTα related with the chromatin activity.
In addition to nuclear activity, studies on the cytoplasmic evolution of ProTα also contributed to deepen the knowledge of its biological function. In this sense, data revealing the natural processing of ProTα to generate Tα1 and Tα11, and those showing the cytoplasmic phosphorylation of ProTα are especially relevant.
The lysosomal asparaginyl endopeptidase, mammalian legumain, is responsible for the processing of ProTα in vivo to generate Tα1 and Tα11, demonstrating the natural origin of these polypeptides23. This research also demonstrates that Tα1 is the main product in the cytoplasm, showing a concentration similar to that of ProTα. Although potential immunotherapeutic effects of Tα1 have been recently reported24, the mechanism of the biological role of this polypeptide remains elusive.
Cytoplasmic phosphorylation of ProTα in threonine residues is dependent on the proliferation activity25,26 and the carcinogenic transformation of the cells. Thus, residues Thr12 or Thr13 are phosphorylated in addition to Thr7 in normal lymphocytes while in carcinogenic lymphocytes Thr7 is the only residue phosphorylated27. The M2 isoenzyme of the pyruvate kinase is the enzyme responsible for the phosphorylation of ProTα that shows this novel dual function28.
A cytoplasmic function of ProTα as an anti-apoptotic factor has also been reported. ProTα seems to be related to the control of the apoptosome activity counteracting the pro-apoptotic action of the protein ANP32A (also known as PHAPI, pp32 or I1PP2A) to promote the caspase-3 activation29. Experiments using cells transformed with ProTα mutated to prevent its phosphorylation, indicate that phosphorylation of ProTα might be required for its anti-apoptotic activity18. This role of ProTα in the apoptotic control is shared by other acidic proteins like SET30 (also named TAF-I, PHAPII or I2PP2A) and ANP32B31 (also named PHAPI2, and APRIL). These proteins, and ANP32A, are nucleus-cytoplasmic shuttling proteins with a long stretch of acidic amino acids. They have a remarkable structural and functional homology with ProTα and a common ability to interact with histones and to influence the chromatin activity. In Figure 2 are shown their sequences.

Figure 2: Comparison of the human sequences of SET, ANP32A, ANP32B and ProTα.
Stretches of acidic amino acid residues are indicated in bold type. Protein sequences were obtained from https://www.ncbi.nlm.nih.gov/protein/.
Although implication of SET in the apoptotic control is well characterized, the mechanisms by which ProTα and the ANP32 proteins are implicated in this process remain elusive. Data from our laboratory, recently published in the Archives of Biochemistry and Biophysics35, reveal interactions of ProTα with SET, ANP32A, ANP32B and other cytoplasmic and mitochondrial proteins that shed some light on the mechanisms of a cytoplasmic action of ProTα and their targets. According to the actual data on the cellular site of action of ProTα, reports showing extracellular effects of this protein should be questioned, at least until the mechanisms for its cellular secretion can be demonstrated.
The interactions of ProTα with SET, ANP32A and ANP32B in lymphocytes
Affinity chromatography and co-immunoprecipitation experiments performed in subcellular fractions of transformed human T-lymphocytes (Jurkat cells) show a novel ability of ProTα to interact in vivo with ANP32A and ANP32B proteins in the cytoplasm of these cells. These results also confirm the ability of ProTα to interact with SET36.
Further experiments with Jurkat cells previously treated with formaldehyde to stabilize possible interactions by chemical cross-linking, lead to isolating six acidic protein complexes ranging from 25 to 45 kDa. These results confirm the ability of ProTα to interact with SET and ANP32 proteins.
The composition of the ProTα-complexes (see Figure 3) denoted a different association of ProTα with SET (complexes 40 kDa and 22 kDa), ANP32A and ANP32B (complexes 30, 28 and 25 kDa), and ANP32A (complex 34 kDa). Other components in complexes of 40, 34, 30 and 25 kDa, are multifunctional proteins which would relate them with cytoplasmic functions such as: proteolytic processing, membrane trafficking, mitochondrial permeability and cell proliferation.
The characterized interactions of ProTα with SET and ANP32 proteins reveal a significant capacity of these acidic proteins to associate. This property has been already stated in complexes in which interactions between SET and ANP32 proteins are involved in the apoptotic control30 or in the regulation of histone acetylation37.
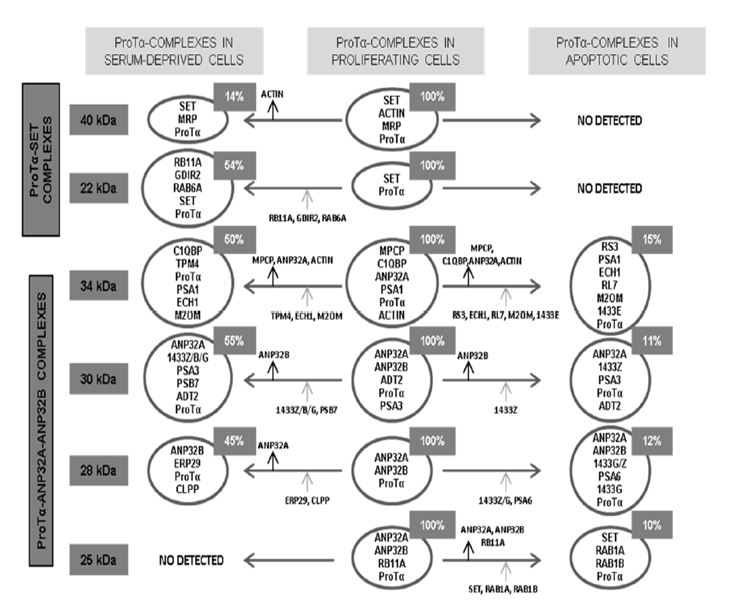
Figure 3: Scheme summarizes the composition of the ProTα-immunoreactive complexes isolated from proliferating Jurkat cells, as well as their evolution when proliferating activity is reduced by serum deprivation or apoptosis induced by staurosporine.
Arrows indicate addition (grey arrows) or loss (black arrows) of proteins in the diverse complexes from proliferating cells when proliferation is reduced or apoptosis induced. Relative abundance of each complex is indicated in the upper insets. Acronyms of proteins other than ProTα, SET, ANP32A, ANP32B and β-actin present in the different complexes: 40 kDa: MRP (MARCKS-related protein); 22 kDa: RB11A (Ras-related protein Rab-11A), GDIR2 (Rho GDP-dissociation inhibitor 2), RAB6A (Ras-related protein Rab-6A); 34 kDa: MPCP (Phosphate carrier protein, mitochondrial), C1QBP (Complement component 1Q subcomponent-binding protein), PSA1 (Proteasome subunit alpha type-1), TPM4 (Tropomyosin alpha-4 chain), ECH1 (Delta(3,5)-delta(2,4)dienoyl-CoA isomerase, mitochondrial), M2OM (Mitochondrial 2- oxoglutarate/malate carrier protein), RS3 (40S ribosomal protein S3), RL7 (60S ribosomal protein L7), 1433 E (14-3-3 protein epsilon); 30 kDa: ADT2 (ADP/ATP translocase 2), PSA3 (Proteasome subunit alpha type-3), PSB7 (Proteasome subunit beta type-7), 1433Z/B/G (14-3-3 protein zeta-delta/beta-alpha/gamma); 28 kDa: ERP29 (Endoplasmatic reticulum resident protein 29), CLPP (Putative ATP-dependent Clp protease proteolytic subunit), PSA6 (Proteasome subunit alpha type-6), 1433G/Z; 25 kDa: RB11A (Rasrelated protein Rab-11A), RAB1A/1B (Ras-related proteins Rab-1A and Rab-1B).
Quantification in the affinity chromatography and co-immunoprecipitation experiments of ProTα, SET and ANP32A and ANP32B proteins, as well as in the isolated complexes, indicate that only a small part of the cellular content of these proteins (about 10%) is implicated in the characterized interactions. This is in agreement with the low cytoplasmic concentration of these proteins found in the immunofluorescence analysis of proliferating lymphocytes35. However, data from the evaluation of the subcellular concentrations of ProTα, SET, ANP32 proteins in extracts from Jurkat cells show a surprising preponderant cytoplasmic presence of these proteins35. This effect might be due to their nuclear leaking produced during the homogenization procedure, as previously demonstrated with ProTα20.
The loss of interaction of ProTα with its targets is observed when cell proliferation of the Jurkat cells is reduced by serum deprivation or apoptosis induced by staurosporine. These effects seem to indicate that these interactions are related to cell proliferation and survival. In Figure 3 is shown the composition of the ProTα-complexes and their evolution in serum-deprived and apoptotic cells.
The cytoplasmic functions attributed to the proteins interacting with ProTα, like ANP32A29 and β-actin38 leads to the hypothesis that ProTα might exert its anti-apoptotic effect counteracting the pro-apoptotic effect described for these proteins. On the contrary, and considering the anti- apoptotic activity of ANP32B31, the interactions with ProTα might reflect its role as a collaborative factor. In addition, interactions of ProTα with SET influencing cell survival might be related to different mechanisms of those attributed to SET as the anti-apoptotic factor through inhibition of the nuclease NM23-H130. On the other hand, interactions of ProTα with ANP32A/ANP32B in complexes 34 and 30 kDa, seem to be involved in mechanisms in which the mitochondrial activity is implicated.
Concluding remarks
Novel interactions described for ProTα provide clues for the understanding of the cytoplasmic activity of ProTα and its targets. The biological functions attributed to proteins interacting with ProTα, lead to hypothesize that the ProTα-complexes are implicated in cell survival by preventing the progress of apoptotic pathways in proliferating lymphocytes. This ability of ProTα supports its role as a multitasker protein with differentiated nuclear and cytoplasmic functions. The scheme in Figure 4 summarizes the cellular processes involving ProTα according to currently available data.
Most of the cellular ProTα migrates to the nucleus of proliferating cells to affect the chromatin activity. Cytoplasmic ProTα undergoes a proteolytic processing to generate Tα1 and a small part of the ProTα pool (about 10%) is phosphorylated by M2-PK concomitantly with cell proliferation. A similar amount of ProTα is involved in the interactions with SET, ANP32A, ANP32B and other cytoplasmic and mitochondrial proteins. On this regard, it should be mentioned the possibility that phosphorylated ProTα could be implicated in such protein interactions as deduced from the analysis of levels of effector caspases in cells transfected with ProTα18. However, we failed to detect ProTα-phosphorylated peptides in the structural analysis of the ProTα-complexes. This question should be addressed in future research to elucidate the functional mechanisms of ProTα and its targets.
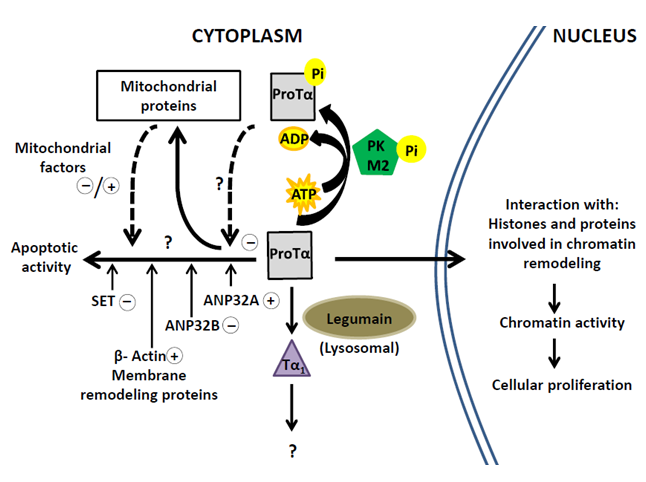
Figure 4: Pathways of Prothymosin α in proliferating cells related to nuclear and cytoplasmic activities through interactions with the indicated proteins.
Questions marks indicate insufficiently characterized mechanisms. Signs +/-, respectively, indicate assigned pro-apoptotic/ anti-apoptotic effects.
Disclosure
The authors have declared that no conflict of interest exists.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Goldstein AL, Slater FD, White A. Preparation, assay and partial purification of a thymic lymphocytopoietic factor (thymosin). Proc Nat Acad Sci USA. 1966; 56(3): 1010-1017.
- Goldstein AL, Guha, A, Zatz M, et al. Purification and biological activity of thymosin, a hormone of the thymus gland. Proc Nat Acad Sci USA. 1972; 69(7): 1800-1803.
- Low TLK, Thurman GB, McAdoo M, et al. The chemistry and biology of thymosin. I. Isolation, characterization, and biological activities of thymosin α1 and polypeptide α1 from calf thymus. J Biol Chem. 1979; 254(3): 981-986.
- Caldarella J, Goodall GJ, Felix AM, et al. Thymosin α11: A peptide related to thymosin α1 isolated from calf thymosin fraction 5. Proc Nat Acad Sci USA. 1983; 80(24): 7424-7427.
- Low TLK, Goldstein AL. Thymic hormones: an overview. In G. D. Sabato, J. J. Langone, & H. V. Vunakis (Eds.), Methods in enzymology. 1985; vol. 116 (pp 213-219). San Diego: Academic Press, Inc.
- Freire M, Crivellaro O, Isaacs C, et al. Translation of mRNA from calf thymus in the wheat germ system: Evidence for a precursor of thymosin α1. Proc Nat Acad Sci USA. 1978; 75(12): 6007-6011.
- Freire M, Hannappel E, Rey M, et al. Purification of thymus mRNA coding for a 16,000 dalton polypeptide containing the thymosin α1 sequence. Proc Nat Acad Sci USA. 1981; 78(1): 192-195.
- Haritos AA, Goodall GJ, Horecker BL. Prothymosin α isolation and properties of the major immunoreactive form of thymosin α1 in rat thymus. Proc Nat Acad Sci USA. 1984; 81(4): 1008-1011.
- Goodall GJ, Dominguez F, Horecker BL. Molecular cloning of cDNA for human prothymosin α. Proc Nat Acad Sci USA. 1986; 83(23): 8926–8928.
- Haritos AA, Tsolas O, Horecker BL. Distribution of prothymosin α in rat tissues. Proc Nat Acad Sci USA. 1984; 81(5): 1391-1393.
- Franco FJ, Díaz C, Barcia M, et al. Thymosin α1 is a native peptide in several tissues. Biochim Biophys Acta. 1992; 1120(1): 43-48.
- Szabo P, Panneerselvam C, Clinton M, et al. Prothymosin alpha gene in humans: organization of its promoter region and localization to chromosome 2. Hum Genet. 1993; 90(6): 629-34.
- Gómez-Márquez J, Segade F, Dosil M, et al. The expression of prothymosin α gene in T lymphocytes and leukemic lymphoid cells is tied to lymphocyte proliferation. J Biol Chem. 1989; 264(15): 8451-8454.
- Bustelo XR, Otero A, Gómez-Márquez J, et al. Expression of the rat prothymosin α gene during T-lymphocyte proliferation and liver regeneration. J Biol Chem. 1991; 266(3): 1443-1447.
- Eschenfeldt WH, Manrow RE, Krug MS, et al. Isolation and partial sequencing of the human prothymosin α gene family. Evidence against export of the gene products. J Biol Chem. 1989; 264(13): 7546-7555.
- Nogueira M, Freire M. Characteristics of the translation of thymosin alpha 1 precursor mRNA by cell-free wheat germ system. Evidence for the acetylation of thymosin alpha 1 precursor. Int J Biochem. 1985; 17(4): 533-536.
- Conteas CN, Mutchnick MG, Palmer KC, et al. Cellular levels of thymosin immunoreactive peptides are linked to proliferative events: evidence for a nuclear site of action. Proc Nat Acad Sci. USA. 1990; 87(9): 3269-3273.
- Moreira D, Díaz-Jullien C, Sarandeses CS, et al. The influence of phosphorylation of prothymosin α on its nuclear import and antiapoptotic activity. Biochem. Cell Biol. 2013; 91: 265-269.
- Manrow RE, Sburlati AR, Hanover JA, et al. Nuclear targeting of prothymosin α. J Biol Chem.1991; 266(6): 3916-3924.
- Covelo G, Sarandeses CS, Díaz-Jullien C, et al. Prothymosin α interacts with free core histones in the nucleus of dividing cells. J Biochem. 2006; 140(5): 627-637.
- Freire J, Covelo G, Sarandeses C, et al. Identification of nuclear-import and cell-cycle regulatory proteins that bind to prothymosin α. Biochem Cell Biol. 2001; 79: 123-131.
- Potaczek DP, Harb H, Michel S, et al. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. 2017; 9(4): 539-571.
- Sarandeses CS, Covelo G, Díaz-Jullien C, et al. Prothymosin α is processed to thymosin α1 and thymosin α11 by a lysosomal asparaginyl endopeptidase. J Biol Chem. 2003; 278(15): 13286-13293.
- Romani L, Oikonomou V, Moretti S, et al. Thymosin α1 represents a potential potent single-molecule-based therapy for cystic fibrosis. Nat Med. 2017; 23: 590–600.
- Barcia MG, Castro JM, Jullien CD, et al. Prothymosin α is phosphorylated in proliferating stimulated cells. J Biol Chem. 1993; 268(7): 4704-4708.
- Pérez-Estévez A, Díaz-Jullien C, Covelo G, et al. A 180-kDa protein kinase seems to be responsible for the phosphorylation of prothymosin α observed in proliferating cells. J Biol Chem. 1997; 272(16): 10506-10513.
- Díaz-Jullien C, Moreira D, Sarandeses CS, et al. The M2-type isoenzyme of pyruvate kinase phosphorylates prothymosin alpha in proliferating lymphocytes. Biochem Biophys Acta. 2011; 1814: 355-365.
- Freire M, Sarandeses CS, Covelo G, et al. Phosphorylation of Prothymosin α. An Approach to Its Biological Significance. In Thymosins (Gerald Litwack, ed.). 2016; vol. 102: pp. 73-99. Academic Press, UK
- Jiang X, Kim HE, Shu H, et al. Distinctive roles of PHAP proteins and prothymosin alpha in a death regulatory pathway. Science. 2003; 299: 223-226.
- Fan Z, Beresford PJ, Oh DY, et al. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003; 112: 659-672.
- Shen SM, Yu Y, Wu YL, et al. Downregulation of ANP32B, a novel substrate of caspase-3, enhances caspase-3 activation and apoptosis induction in myeloid leukemic cells. Carcinogenesis. 2010; 31: 419-426.
- Matilla A, Radrizzani M. The Anp32 family of proteins containing leucine-rich repeats. Cerebellum. 2005; 4: 7-18.
- Roth W, Wagenknecht B, Klumpp A, et al. APRIL, a new member of the tumor necrosis factor family, modulates death ligand-induced apoptosis. Cell Death Differ. 2001; 8: 403-410.
- Adachi Y, Pavlakis GN, Copeland TD. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakproint in acute undifferentiated leukemia. FEBS Lett. 1994; 340: 231-235.
- Barbeito P, Sarandeses CS, Díaz-Jullien C, et al. Prothymosin α interacts with SET, ANP32A and ANP32B and other cytoplasmic and mitochondrial proteins in proliferating cells. Arch Biochem Biophys. 2017; 635: 74-86.
- Karetsou Z, Martic G, Tavoulari S, et al. Prothymosin α associates with the oncoprotein SET and is involved in chromatin decondensation. FEBS Lett. 2004; 577: 496-500.
- Seo SB, McNamara P, Heo S, et al. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001; 104: 119-130.
- Tang HL, Le AHP, Lung HL. The increase in mitochondrial association with actin precedes Bax translocation in apoptosis. Biochem J. 2006; 396: 1-5.
