Association of Dementia Human Leukocyte Antigen (HLA) Profile with Human Herpes Viruses 3 and 7: An in-silico Investigation
Lisa M. James1,2,3, Spyros A. Charonis1,2, Apostolos P. Georgopoulos1,2,3,4*
1The HLA Research Group, Brain Sciences Center, Department of Veterans Affairs Health Care System, Minneapolis, MN, 55417, USA
2Department of Neuroscience, University of Minnesota Medical School, Minneapolis, MN 55455, USA
3Department of Psychiatry, University of Minnesota Medical School, Minneapolis, MN 55455, USA
4Department of Neurology, University of Minnesota Medical School, Minneapolis, MN 55455, USA
Abstract
Human leukocyte antigen (HLA), the most highly polymorphic region of the human genome, is increasingly recognized as an important genetic contributor to dementia risk and resilience. HLA is involved in protection against foreign antigens including human herpes viruses (HHV), which have been widely implicated in dementia. Here we used an in silico approach1 to determine binding affinities of glycoproteins from 9 human herpes virus (HHV) strains to 113 HLA alleles, and to examine the association of a previously identified HLA-dementia risk profile2 to those affinities. We found a highly significant correlation between high binding affinities of HLA alleles to HHV 3 and 7 and the dementia risk scores of those alleles, such that the higher the estimated binding affinity, the lower the dementia risk score. These findings suggest that protection conferred by HLA alleles may be related to their ability to bind and eliminate HHV3 and HHV7 and point to the possibility that protection against these viruses may reduce dementia incidence.
Introduction
The human leukocyte antigen (HLA) region is increasingly recognized as an important genetic contributor to dementia risk and resilience2-10. HLA plays a central role in host protection against foreign antigens such as those derived from pathogens including viruses and bacteria. HLA protection against such pathogens is facilitated by two main classes of HLA genes that code for cell-surface proteins that are instrumental in immune system responses to foreign antigens. Class I HLA molecules (HLA-A, B, and C genes) present intracellular antigen peptides to CD8+ cytotoxic T cells to signal destruction of infected cells whereas Class II HLA molecules (HLA-DR, DQ, and DP genes) present endocytosed extracellular antigen peptides to CD4+ T cells to promote B-cell mediated antibody production and immune memory. These two classes of HLA work in concert to facilitate pathogen elimination.
The effectiveness of HLA in pathogen elimination, however, hinges in part on the binding affinity between HLA proteins with those derived from a given pathogen. The HLA region is the most highly polymorphic in the human genome and subtle variations in HLA proteins have been shown to affect binding affinity thereby influencing disease outcomes11,12. Indeed, HLA has been implicated in more diseases than any other region of the human genome13 and is particularly implicated in autoimmune conditions14, and increasingly, in dementia2-10. With regard to dementia, we have used a population immunogenetic epidemiological approach in 14 Continental Western European countries to identify an HLA-dementia risk profile characterized by 127 HLA alleles that are either negatively correlated with the population prevalence of dementia and are therefore considered protective or HLA alleles that are positively correlated with the population prevalence of dementia and are presumed to promote susceptibility2. We hypothesized that HLA protection against dementia is related to superior binding affinity to harmful antigens and that, conversely, HLA susceptibility to dementia is related to persistent foreign antigens (due to poor binding affinity) that may directly damage cells and/or stimulate chronic inflammatory responses and autoimmunity2.
Numerous pathogens have been implicated in dementia15,16, perhaps none more so than human herpes viruses (HHV)17-22. Although HHV are nearly ubiquitous23-25, dementia is relatively rare; we have previously suggested that HLA variations may moderate the association of HHV infection and dementia such that some alleles may provide superior binding affinity to specific strains, thus exerting a protective role for dementia1. In an initial in silico investigation, we evaluated the binding affinity of three dementia-protective and three neutral Class II HLA alleles to HHV epitopes1. We found that the HLA alleles that were protective against dementia had significantly higher binding affinity to HHV epitopes than the neutral alleles, with the most significant differences found for HHV6 glycoproteins. Here we extend that line of research to evaluate the association between the dementia risk scores of a large number of HLA Class I and II alleles previously characterized in an HLA-dementia risk profile2 and the affinity of those alleles with glycoproteins from 9 HHV strains.
Materials and Methods
HHV proteins
The amino acid sequences of surface glycoproteins from HHV 1-8 (1, 2, 3, 4, 5, 6A, 6B, 7, 8) were retrieved from the UniprotKB database26. Table 1 gives the details of these proteins and associated information regarding the number of n-mers used in the analyses of Class I and Class II alleles.
Table 1: HHV proteins used. See text for details.
|
Virus |
Protein description |
UniprotKB ID |
Protein length (AA) |
|
HHV-1 |
Envelope glycoprotein D |
Q69091 |
394 |
|
HHV-2 |
Envelope glycoprotein D |
P03172 |
393 |
|
HHV-3 |
Envelope glycoprotein E |
Q9J3M8 |
623 |
|
HHV-4 |
Envelope glycoprotein B |
P03188 |
857 |
|
HHV-5 |
Envelope glycoprotein B |
P06473 |
906 |
|
HHV-6A |
Glycoprotein Q2 |
P0DOE0 |
214 |
|
HHV-6B |
Glycoprotein Q1 |
Q9QJ11 |
516 |
|
HHV-7 |
Envelope glycoprotein H |
P52353 |
690 |
|
HHV-8 |
Envelope glycoprotein H |
F5HAK9 |
730 |
A sliding window approach1 was used to partition the sequence of each glycoprotein into subsequences of 9-mers (for HLA Class I analyses; Fig. 1) and 22-mers (for HLA Class II analyses; Fig. 2) that covered the entire length of the protein. These n-mers were chosen given that they are typically short for Class I molecules (8-10 amino acids, AA) and around 20 for Class II molecules27. For each n-mer, a set of subsequences was generated (number of subsequences = length of glycoprotein – n). For both kinds of n-mers (n=9, Class I, and n=22, Class II), subsequences were collected and queried in the IEDB database (www.iedb.org) in order to identify potential epitope peptides that are recognized by and bind to HLA Class I and II surface receptor proteins. IEDB queries were performed for each of the sliding-window aggregated sequences against 113 HLA alleles (55 Class I and 58 Class II; see below). Binding affinity predictions were obtained using the NetMHCIIpan method28. For each n-mer, a binding affinity score was predicted and reported as a percentile rank by comparing the peptide’s score against the scores of five million random n-mers selected from the SwissProt database26; smaller percentile ranks indicate higher binding affinity (“good binders”). For each HHV strain and HLA allele, the average percentile rank of n-mer ranks <1 (APR<1) was used as a high binding affinity measure for quantitative analyses.

Figure 1: The sliding-window testing method is illustrated for 9-mers, as applied to the envelope glycoprotein H amino acid sequence of HHV7 to investigate binding affinities to HLA Class I molecules. Capital case letters in the sequence denote a specific amino acid residue. Brown horizontal lines indicate four 9-mer subsequences obtained by shifting the sliding window by one amino acid residue at a time.
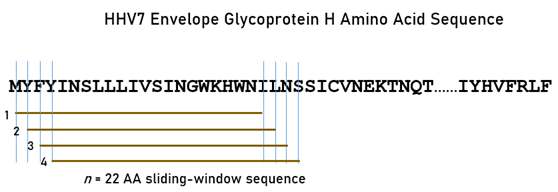
Figure 2. The sliding-window testing method is illustrated for 22-mers, as applied to the envelope glycoprotein H amino acid sequence of HHV7 to investigate binding affinities to HLA Class II molecules. Capital case letters in the sequence denote a specific amino acid residue. Brown horizontal lines indicate four 22-mer subsequences obtained by shifting the sliding window by one amino acid residue at a time.
HLA alleles
We used 113 HLA alleles out of 127 alleles for which we had obtained a dementia-HLA profile2 using data from 14 Continental Western European Countries (Austria, Belgium, Denmark, Finland, France, Germany, Greece, Italy, Netherlands, Portugal, Norway, Spain, Sweden, and Switzerland). Those 127 alleles comprised 69 Class I and 58 Class II alleles2; of those, in the present study we used all 58 Class II alleles (N = 15, 14, 29 for DPB1, DQB1 and DRB1 genes, respectively) but only 55 Class I alleles (N = 17, 25, 13 for A, B, and C genes, respectively) because 14 alleles could not be modeled by the NetMHCIIpan method.
Dementia-HLA profile (dementia risk scores)
The dementia-HLA profile consisted of 113 Fisher z-transformed correlations r´ between each HLA allele frequency and log-transformed dementia prevalence2. These values can be regarded as continuous-varying HLA-related dementia-risk scores, such that (a) their sign indicates protection (negative) or susceptibility (positive) to dementia, and (b) their absolute value indicates the strength of their effect (protection or susceptibility). Of the 113 alleles here, 51 were dementia-protective alleles (negative scores) and 62 dementia-susceptibility alleles (positive scores).
Data analysis
The main objective of this study was to evaluate the association between specific HLA-HHV strain binding affinity and the HLA-dementia risk scores. For this purpose, we computed the Pearson correlation coefficient between the dementia risk scores and HHV-specific HLA affinity (APR<1) for each HHV strain. For the group of alleles with APR ≥ 1, i.e. alleles that were not used in the correlation analysis above, we tested the proportion of susceptibility alleles against the null hypothesis of the proportion = 0.5 using a one-sample binomial test of proportions. The IBM-SPSS statistical package (version 27) was used for these analyses.
Results
The frequency distributions of the HLA-dementia risk scores and APR<1 values (all HHV strains) are shown in Figures 3 and 4, respectively. For each HHV strain, the dementia risk scores of individual alleles are plotted against the APR<1 value of the corresponding allele in Figure 5. The correlation coefficients and associated statistics for each HHV strain are given in Table 3. Statistically significant correlations were found only for HHV3 (blue in Fig. 5) and HHV7 (red in Fig. 5). The pooled APR<1 values for these two HHV strains are plotted in Figure 6; the Pearson correlation coefficient was 0.270 (P = 0.00029, N = 176).
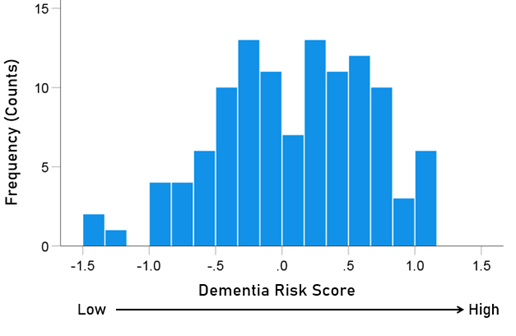
Figure 3. Frequency distribution of the dementia risk scores of the 113 alleles used (Table 2).
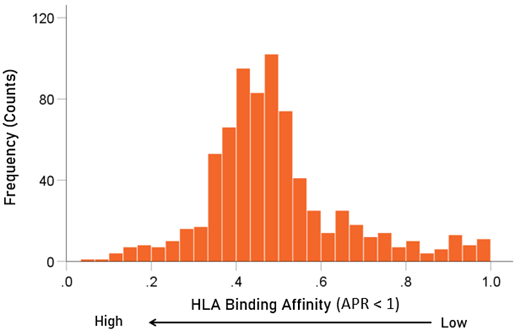
Figure 4. Frequency distribution of APR<1 values (N = 752 of a total of 9 HHV strains x 113 HLA alleles = 107 values; the remainder had values of APR≥1).
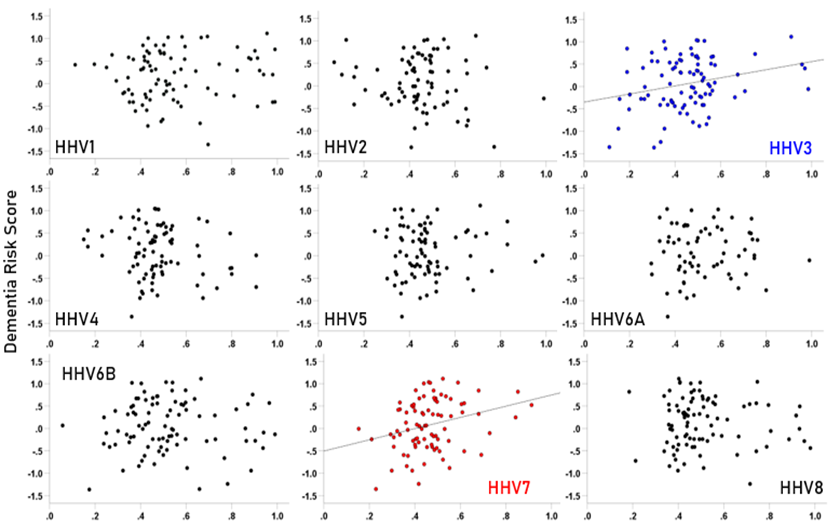
Figure 5. Scatter plots of dementia risk scores against APR<1 values for the each HHV strain. A statistically significant correlation was found only for HHV3 (r = 0.260, P = 0.013, N = 91) and HHV7 (r = 0.287, P = 0.008, N = 85). All correlations and associated statistics are given in Table 3.
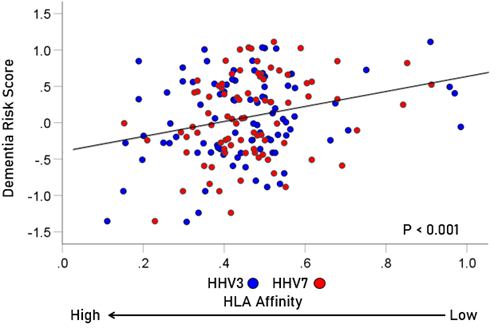
Figure 6. Scatter plot of dementia risk scores against APR<1 values for the pooled data of HHV3 and HHV7 (r = 0.270, P = 0.00029, N = 176).
Table 2: Dementia-risk scores ()2 of the 113 HLA alleles used. See text for details.
|
|
Allele |
Dementia risk scores (r´) |
|
1 |
A*01:01 |
−0.237 |
|
2 |
A*02:01 |
−0.593 |
|
3 |
A*02:05 |
0.853 |
|
4 |
A*03:01 |
−0.412 |
|
5 |
A*11:01 |
0.347 |
|
6 |
A*23:01 |
0.317 |
|
7 |
A*24:02 |
0.051 |
|
8 |
A*25:01 |
−0.031 |
|
9 |
A*26:01 |
0.478 |
|
10 |
A*29:02 |
0.131 |
|
13 |
A*31:01 |
0.357 |
|
14 |
A*32:01 |
0.580 |
|
15 |
A*33:01 |
−0.361 |
|
16 |
A*68:01 |
0.392 |
|
17 |
A*68:02 |
0.672 |
|
11 |
A*30:01 |
−0.220 |
|
12 |
A*30:02 |
−0.137 |
|
18 |
B*07:02 |
−0.941 |
|
19 |
B*08:01 |
−0.797 |
|
20 |
B*13:02 |
−0.240 |
|
21 |
B*14:02 |
0.719 |
|
22 |
B*15:01 |
−0.611 |
|
23 |
B*18:01 |
0.597 |
|
24 |
B*27:02 |
0.060 |
|
25 |
B*27:05 |
−0.195 |
|
26 |
B*35:01 |
0.306 |
|
27 |
B*35:03 |
0.672 |
|
28 |
B*37:01 |
−0.412 |
|
29 |
B*38:01 |
1.026 |
|
30 |
B*39:01 |
0.284 |
|
31 |
B*40:01 |
−0.509 |
|
32 |
B*40:02 |
−0.010 |
|
33 |
B*44:02 |
−0.841 |
|
34 |
B*44:03 |
0.086 |
|
35 |
B*49:01 |
0.847 |
|
36 |
B*50:01 |
0.509 |
|
37 |
B*51:01 |
0.535 |
|
38 |
B*52:01 |
0.317 |
|
39 |
B*55:01 |
−0.479 |
|
40 |
B*56:01 |
−0.309 |
|
41 |
B*57:01 |
−0.166 |
|
42 |
B*58:01 |
0.721 |
|
43 |
C*01:02 |
−0.011 |
|
44 |
C*03:03 |
−0.416 |
|
45 |
C*04:01 |
1.035 |
|
46 |
C*05:01 |
−0.174 |
|
47 |
C*06:02 |
−0.135 |
|
48 |
C*07:01 |
−0.362 |
|
49 |
C*07:02 |
−0.883 |
|
50 |
C*07:04 |
−0.238 |
|
51 |
C*12:02 |
0.636 |
|
52 |
C*12:03 |
1.007 |
|
53 |
C*14:02 |
0.681 |
|
54 |
C*15:02 |
0.660 |
|
55 |
C*16:01 |
0.275 |
|
56 |
DPB1*01:01 |
−0.941 |
|
57 |
DPB1*02:01 |
1.112 |
|
58 |
DPB1*02:02 |
0.405 |
|
59 |
DPB1*03:01 |
0.493 |
|
60 |
DPB1*04:01 |
−1.238 |
|
61 |
DPB1*04:02 |
−0.438 |
|
62 |
DPB1*05:01 |
−0.408 |
|
63 |
DPB1*06:01 |
0.199 |
|
64 |
DPB1*09:01 |
1.043 |
|
65 |
DPB1*10:01 |
0.809 |
|
66 |
DPB1*11:01 |
0.007 |
|
67 |
DPB1*13:01 |
0.291 |
|
68 |
DPB1*14:01 |
0.223 |
|
69 |
DPB1*17:01 |
0.429 |
|
70 |
DPB1*19:01 |
−0.241 |
|
71 |
DQB1*02:01 |
−0.413 |
|
72 |
DQB1*02:02 |
0.197 |
|
73 |
DQB1*03:01 |
0.569 |
|
74 |
DQB1*03:02 |
−0.280 |
|
75 |
DQB1*03:03 |
−0.282 |
|
76 |
DQB1*04:02 |
−0.091 |
|
77 |
DQB1*05:01 |
−0.275 |
|
78 |
DQB1*05:02 |
0.560 |
|
79 |
DQB1*05:03 |
0.428 |
|
80 |
DQB1*06:01 |
0.267 |
|
81 |
DQB1*06:02 |
−0.696 |
|
82 |
DQB1*06:03 |
−0.767 |
|
83 |
DQB1*06:04 |
−0.662 |
|
84 |
DQB1*06:09 |
0.001 |
|
85 |
DRB1*01:01 |
−0.722 |
|
86 |
DRB1*01:02 |
0.540 |
|
87 |
DRB1*01:03 |
0.067 |
|
88 |
DRB1*03:01 |
0.494 |
|
89 |
DRB1*04:01 |
−1.363 |
|
90 |
DRB1*04:02 |
0.758 |
|
91 |
DRB1*04:03 |
0.524 |
|
92 |
DRB1*04:04 |
−0.278 |
|
93 |
DRB1*04:05 |
1.020 |
|
94 |
DRB1*04:07 |
0.248 |
|
95 |
DRB1*04:08 |
−0.341 |
|
96 |
DRB1*07:01 |
−0.007 |
|
97 |
DRB1*08:01 |
−0.104 |
|
98 |
DRB1*08:03 |
−0.057 |
|
99 |
DRB1*09:01 |
−0.131 |
|
100 |
DRB1*10:01 |
0.288 |
|
101 |
DRB1*11:01 |
0.847 |
|
102 |
DRB1*11:02 |
0.564 |
|
103 |
DRB1*11:03 |
0.820 |
|
104 |
DRB1*11:04 |
0.726 |
|
105 |
DRB1*12:01 |
−0.180 |
|
106 |
DRB1*13:01 |
−0.509 |
|
107 |
DRB1*13:02 |
−0.589 |
|
108 |
DRB1*13:03 |
0.821 |
|
109 |
DRB1*13:05 |
0.324 |
|
110 |
DRB1*14:01 |
0.361 |
|
111 |
DRB1*15:01 |
−1.353 |
|
112 |
DRB1*15:02 |
0.416 |
|
113 |
DRB1*16:01 |
0.544 |
Table 3: Pearson correlation coefficients between dementia risk scores and APR<1 values for the 9 HHV strains.
|
HHV Strain |
Correlation coefficient |
P value |
N |
|
1 |
0.063 |
0.567 |
85 |
|
2 |
−0.131 |
0.248 |
79 |
|
3 |
0.260** |
0.013 |
91 |
|
4 |
−0.172 |
0.120 |
83 |
|
5 |
0.031 |
0.780 |
83 |
|
6A |
0.025 |
0.840 |
67 |
|
6B |
−0.007 |
0.951 |
89 |
|
7 |
0.287*** |
0.008 |
85 |
|
8 |
−0.038 |
0.719 |
90 |
For HHV3 and HHV7, there were a total of 2 x 113 = 226 alleles tested. Of those, 176 had values of APR < 1, and, hence were used for the correlation analysis above (Fig. 6). The remaining 50 alleles had values of APR ≥ 1 and were not eligible for the correlation analysis. However, we were interested to find out whether that group was enriched with susceptibility (positive) dementia risk scores, a hypothesis in line with the correlation findings above. Indeed, the proportion of susceptibility scores in that group was 33/50 = 66%, a proportion significantly higher than that of the null hypothesis of 50% (binomial one-sample proportions test, z = 2.263, two-sided P = 0.024).
Discussion
In this in silico study we evaluated estimated binding affinities of glycroproteins from 9 HHV strains and 113 HLA alleles and examined the association of those affinities with a previously-identified HLA-dementia risk profile2. The findings indicated that for HHV3 and HHV7, but not for other HHV, the glycoprotein binding affinities were significantly associated with the dementia risk score such that higher binding affinity was associated with lower dementia risk. These findings suggest that protection conferred by dementia-protective HLA alleles is related, in part, to their ability to bind and eliminate HHV3 and HHV7. These findings add to the literature linking HHV to dementia and implicate HHV3 and HHV7, in particular, in dementia risk.
HHV3, often referred to as varicella-zoster virus (VZV), is a highly transmissible disease that results in chicken pox (varicella) upon initial infection, typically in childhood, and after a period of latency may reactivate as shingles (herpes zoster) in later adulthood. Acute varicella infection is usually mild, however, reactivation as herpes zoster is associated with several neurological complications including stroke, postherpetic neuralgia, meningitis, myelopathy, and ocular disorders29-31. Moreover, VZV infection has been associated with cognitive decline32 and dementia33-36. Among those infected, antiviral treatment has been associated with reduced dementia risk33-36, providing further evidence supporting a role of HHV3 in dementia pathogenesis. HHV3 is the only herpes virus for which effective vaccines exist – one to prevent varicella and another to prevent herpes zoster31. Introduction of vaccination programs has resulted in a rapid decrease in varicella37 and herpes zoster38 incidence. Furthermore, preliminary evidence indicates that shingles vaccination is associated with reduced incidence of dementia39.
HHV7 is an extremely common virus that affects most humans primarily during childhood with HHV7 seroprevalence reaching 76% in children aged 3-640 and 98% in adults41. HHV7, which is commonly referred to as Roseolovirus along with genetically and biologically similar HHV6 variants42,43, is predominantly transmitted in saliva and is continuously shed in saliva of healthy adults44. Infection with HHV7 has been shown to result in cytopathic effects43 44, down regulation of CD4+ cells45,46, and altered trafficking of Class I MHC molecules47. Like other herpesviruses, HHV7 persists after the primary infection as a lytic or latent infection that can be reactivated, or can reactivate other HHV48, contributing to disease42-44. HHV7 has most widely been associated with exanthem subitum and pityriasis rosea44. In addition, HHV7 has been associated with neurological signs and symptoms49 and with demyelinating diseases50, and recently, with dementia51. Specifically, increased HHV7 was found in post-mortem brain tissue of patients with AD relative to controls, and HHV7 corresponded with hallmark AD-related neuropathological findings including amyloid beta deposition51. Notably, recent evidence suggests amyloid beta may be involved in innate immunity, exerting antimicrobial properties to HHV52.
The findings of the present in silico study provide novel and compelling evidence linking HHV3 and HHV7 to dementia. Considering the present findings in light of the discordant rates of HHV3 and HHV7 infection with dementia prevalence, we suggest that HHV3 and HHV7 may contribute to dementia and associated neuropathology in those lacking HLA alleles that are able to bind with sufficient affinity and immunogenicity to eliminate the virus. In the absence of immunogenetic protection against these viruses, prevention of HHV infection via vaccination may be of paramount importance for reducing dementia incidence. Indeed, previous studies have reported reduced dementia incidence following vaccination for influenza53,54 and tetanus, diphtheria, pertussis (Tdap)55. Preliminary evidence suggests a protective role of shingles vaccination against dementia39 though additional studies are needed to verify those initial intriguing findings. Approved vaccines do not yet exist for other HHV though several are under development, particularly for herpes simplex virus56. The present findings point to a potential benefit of HHV7 vaccine development in reducing dementia risk.
These novel findings, however, must also be considered in terms of several qualifications. First, the HLA-dementia profile at the center of these analyses were derived from Continental Western European countries and may not generalize to other regions given ethnic and geographic variation in HLA57-59. Future studies evaluating the associations of HLA-HHV affinity with HLA-dementia profiles in other populations will be important for corroborating and/or extending the present findings to other HHV strains and populations. Second, the present study only focused on HHV. Numerous microbes and viruses have been implicated in risk for dementia15,16. The same in silico approach used here can readily be applied to other infectious agents to evaluate their binding affinity with HLA alleles associated with dementia susceptibility and protection. Third, while this study identified HHV3 and HHV7 as playing a central role in HLA-mediated dementia, other non-HLA mechanisms that were not investigated here may influence the association of other HHV with dementia. Finally, we exclusively evaluated HLA-HHV glycoprotein binding affinity; binding affinity is one of many factors that contributes to disease outcomes. Additional in vitro and in vivo studies are warranted to further validate the findings of this in silico study and determine the relative influence of HLA-HHV binding affinity on dementia.
In summary, findings of the present study suggest that dementia risk scores, which are characterized by the sign and strength of the correlation between the population frequency of HLA alleles and the population prevalence of dementia, are significantly associated with HLA binding affinity to glycoproteins for HHV3 and HHV7. HLA alleles that bind with strong affinity to HHV3 and HHV7 presumably promote virus elimination, reducing deleterious downstream effects that may lead to dementia.
Acknowledgments
Partial funding for this study was provided by the University of Minnesota (the Anita Kunin Chair in Women's Healthy Brain Aging, the Brain and Genomics Fund, the McKnight Presidential Chair of Cognitive Neuroscience, and the American Legion Brain Sciences Chair). The sponsors had no role in the current study design, analysis or interpretation, or in the writing of this paper. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Author contributions
LMJ and APG conceived the research; SAC carried out the HHV protein retrieval, IEDB query and data preprocessing; APG contributed to data analysis; LMJ, APG and SAC contributed to writing the manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Charonis S, James LM, Georgopoulos AP. In silico assessment of binding affinities of three dementia-protective Human Leukocyte Antigen (HLA) alleles to nine human herpes virus antigens. Curr Res Transl Med. 2020; 68(4): 211-6.
- James LM, Georgopoulos AP. Immunogenetic epidemiology of dementia and Parkinson’s Disease in 14 Continental European countries: Shared human leukocyte antigen (HLA) profiles. J Immunological Sci. 2021; 5(2): 16-26.
- Kunkle BW, Grenier-Boley B, Sims R, et al.Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019; 51: 414–430.
- Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013; 45(12): 1452-1458.
- Steele NZ, Carr JS, Bonham LW, et al. Fine-mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: a case– control study. PLOS Med. 2017; 14(3): e1002272.
- Wang ZX, Wan Q, Xing A. HLA in Alzheimer’s Disease: Genetic association and possible pathogenic roles. Neuromol Med. 2020;22: 464–473.
- Aliseychik MP, Andreeva TV, Rogaev EI. Immunogenetic factors of neurodegenerative diseases: The role of HLA class II. Biochemistry Moscow. 2018;83: 1104–1116.
- James LM, Georgopoulos AP. The human leukocyte antigen (HLA) DRB1*13:02 allele protects against dementia in continental Western Europe. J Neurol Neuromed. 2019; 4(5): 1-6.
- James LM, Georgopoulos AP. Tri-allelic human leukocyte antigen (HLA) protection against dementia. J Neurol Neuromed. 2020; 5(1): 12-17.
- James LM, Georgopoulos AP. Shared human leukocyte antigen (HLA) coverage in dementia and Parkinson’s disease. J Neurol Neuromed. 2020; 5(3): 45-54.
- Hov JR, Kosmoliaptsis V, Traherne JA, et al. Electrostatic modifications of the HLA-DR P9 peptide-binding pocket and susceptibility to primary sclerosing cholangitis. Hepatology. 2011; 53: 1967–
- Davenport MP, Quinn CL, Chicz RM, et al. Naturally processed peptides from two disease-resistance-associated HLA-DR13 alleles show related sequence motifs and the effects of the dimorphism at position 86 of the HLA-DR beta chain. PNAS 1995; 92 (14): 6567–6571.
- Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Ann Rev Genom Hum Genet.2013; 14: 301–323.
- Simmonds MJ, Gough SC. The HLA region and autoimmune disease: Associations and mechanisms of action. Curr Genomics. 2007; 8(7): 453-65.
- Maheshwari P, Eslick GD. Bacterial infection and Alzheimer’s disease: A meta-analysis. J Alzheimers Dis 2015; 43: 957–66.
- Mawanda F, Wallace R. Can infections cause Alzheimer's disease? Epidemiol Rev 2013; 35: 161–
- Itzhaki RF, Lathe R, Balin BJ, et al. Microbes and Alzheimer’s disease. J Alzheimers Dis 2016; 51: 979–84.
- Itzhaki RF. Corroboration of a major role for herpes simplex virus type 1 in alzheimer’s disease. Front Aging Neurosci. 2018; 10: e00324.
- Chen VC, Wu SI, Huang KY, et al. Herpes zoster and dementia: a nationwide population-based cohort study. J Clin Psychiat. 2018; 79: 16m11312.
- Eimer WA, Vijaya Kumar DK, Navalpur Shanmugam NK, et al. Alzheimer’s disease associated beta-amyloid is rapidly seeded by Herpesviridae to protect against brain infection. Neuron. 2018; 99(1): 56–63.
- Readhead B, Haure-Mirande JV, Funk CC, et al. Multiscale analysis of independent Alzheimer’s cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron. 2018; 99(1): 64–82.
- Westman G, Blomberg J, Yun Z, et al. Decreased HHV-6 IgG in Alzheimer’s disease. Front Neurol. 2017; 8: 40.
- McQuillan G, Kruszon-Moran D, Flagg EW, et al. Prevalence of herpes simplex virus type 1 and type 2 in persons aged 14–49: United States, 2015–2016. NCHS Data Brief, no 304. Hyattsville, MD: National Center for Health Statistics. 2018.
- Staras A, Dollard SC, Radford KW, et al. Seroprevalence of cytomegalovirus infection in the United States, 1988– 1994. Clin Infect Dis 2006; 43: 1143–51.
- Wald A, Corey L, et al. Persistence in the population: epidemiology, transmission. In: Arvin A, Campadelli-Fiume G, Mocarski E, editors. Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge: Cambridge University Press; 2007 Chapter 36.
- The Uniprot Consortium 2019 (https://www.uniprot.org/).
- Janice S, Blum JS, Wearsch PA, et al. Pathways of Antigen Processing. Annu Rev Immunol. 2013; 31: 443–73.
- Reynisson B, Alvarez B, Paul S, et al. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020; 48, W449-W454.
- Breuer J, Pacou M, Gautier A, et al. Herpes zoster as a risk factor for stroke and TIA: a retrospective cohort study in the UK. Neurology. 2014; 83(2): e27-33.
- Gilden D, Mahalingam R, Nagel MA, et al. Review: The neurobiology of varicella zoster virus infection. Neuropathol Appl Neurobiol. 2011; 37(5): 441-63.
- Gershon A, Breuer J, Cohen J, et al. Varicella zoster virus infection. Nat Rev Dis Primers. 2015; 1: 1-18.
- Grahn A, Nilsson S, Nordlund A, et al. Cognitive impairment 3 years after neurological Varicella-zoster virus infection: a long-term case control study. J Neurol. 2013; 260: 2761-2769.
- Chen VC, Wu SI, Huang KY, et al. Herpes zoster and dementia: a nationwide population-based cohort study. J Clin Psychiat. 2017; 78(1): 16m11312.
- Tsai MC, Cheng WL, Sheu JJ, et al. Increased risk of dementia following herpes zoster ophthalmicus. PloS One. 2017;12: e0188490.
- Bae S, Yun SC, Kim MC, et al. Association of herpes zoster with dementia and effect of antiviral therapy on dementia: a population-based cohort study. Eur Arch Psychiatry Clin Neurosci. 2021; 271(5): 987-997.
- Lopatko Lindman K, Hemmingsson E-S, Weidung B, et al. Herpesvirus infections, antiviral treatment, and the risk of dementia—a registry-based cohort study in Sweden. Alzheimer's Dement. 2021; 7: e12119.
- Helmuth IG, Poulsen A, Suppli CH, et al. Varicella in Europe-A review of the epidemiology and experience with vaccination. Vaccine. 2015; 33(21): 2406-13.
- James SF, Chahine EB, Sucher AJ, et al. Shingrix: The new adjuvanted recombinant herpes zoster vaccine. Ann Pharmacother. 2018; 52(7): 673-680.
- Schnier C, Janbek J, Lathe R, et al. Reduced dementia incidence following varicella zoster vaccination in Wales 2013–2020. medRxiv. 2021. doi: 10.1101/2021.07.22.21260981
- Cermelli C, Fabio G, Montorsi M, et al. Prevalence of antibodies to human herpesviruses 6 and 7 in early infancy and age at primary infection. New Microbiol 1996; 19: 1–8.
- Krueger GR, Koch B, Leyssens N, et al. Comparison of seroprevalences of human herpesvirus-6 and -7 in healthy blood donors from nine countries. Vox Sang. 1998; 75: 193–7
- Dewhurst S, Skrincosky D, van Loon N. Human herpesvirus 7. Expert Rev Mol Med. 1997; 1(2): 1-10.
- Wang FZ, Pellett PE. HHV-6A, 6B, and 7: immunobiology and host response. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. Chapter 48.
- Black JB, Pellett PE. Human herpesvirus 7. Rev Med Virol. 1999; 9(4): 245-62.
- Furukawa M, Yasukawa M, Yakushijin Y, et al. Distinct effects of human herpesvirus 6 and human herpesvirus 7 on surface molecule expression and function of CD4+ T cells. J. Immunol. 1994; 152: 5768–5775.
- Secchiero P, Gibellini D, Flamand L, et al. Human herpesvirus 7 induces the down-regulation of CD4 antigen in lymphoid T cells without affecting p56lck levels. J. Immunol. 1997; 159: 3412–3423.
- Glosson NL, Gonyo P, May NA, et al. Insight into the mechanism of human herpesvirus 7 U21-mediated diversion of class I MHC molecules to lysosomes. J Biol Chem. 2010; 285(47): 37016-29.
- TanakaâTaya K, Kondo T, Nakagawa N, et al. Reactivation of human herpesvirus 6 by infection of human herpesvirus 7. J Med Virol. 2000; 60(3): 284-9.
- Yoshikawa T, Ihira M, Suzuki K, et al. Invasion by human herpesvirus 6 and human herpesvirus 7 of the central nervous system in patients with neurological signs and symptoms. Arch Dis Child. 2000; 83(2): 170-1.
- Tomsone V, Logina I, Millers A, et al.Association of human herpesvirus 6 and human herpesvirus 7 with demyelinating diseases of the nervous system. J Neurovirol. 2001; 7: 564–569.
- Readhead B, Haure-Mirande JV, Funk CC, et al. Multiscale Analysis of Independent Alzheimer's Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron. 2018; 99(1): 64–82.
- Eimer WA, Kumar DK, Shanmugam NK, et al. Alzheimer’s disease-associated β-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron. 2018; 99(1): 56-63.
- Luo CS, Chi CC, Fang YA, et al. Influenza vaccination reduces dementia in patients with chronic obstructive pulmonary disease: a nationwide cohort study. J Investig Med. 2020; 68: 838–845.
- Liu JC, Hsu YP, Kao PF, et al. Influenza vaccination reduces dementia risk in chronic kidney disease patients: a population-based cohort study. Medicine. 2016; 95: e2868.
- Scherrer JF, Salas J, Wiemken TL, et al. Lower risk for dementia following adult tetanus, diphtheria, and pertussis (Tdap) vaccination. J Gerontol Ser A. 2021; 76(8): 1436–1443.
- Johnston C, Gottlieb SL, Wald A. Status of vaccine research and development of vaccines for herpes simplex virus. Vaccine. 2016; 34(26): 2948-52.
- Buhler S, Sanchez-Mazas A. HLA DNA sequence variation among human populations: molecular signatures of demographic and selective events. PloS one. 2011; 6(2): e14643.
- Sanchez-Mazas A, Lemaître JF, Currat M. Distinct evolutionary strategies of human leucocyte antigen loci in pathogen-rich environments. Philos Trans R Soc Lond B Biol Sci. 2012; 367(1590): 830-9.
- Salamon H, Klitz W, Easteal S, et al. Evolution of HLA class II molecules: Allelic and amino acid site variability across populations. Genetics. 1999; 152(1): 393-400.
