Clinical implications of Epstein-Barr virus strain diversity
Ana Cirac1,2,3, Uta Behrends1,2,3, and Josef Mautner1,2,3*
1Children’s Hospital, Technische Universität München, Munich, Germany
2Research Unit Gene Vectors, Helmholtz Zentrum München, Munich, Germany
2German Centre for Infection Research (DZIF), partner site Munich, Germany
Abstract
The Epstein-Barr virus (EBV), a ubiquitous γ-herpesvirus, has been implicated in the etiology of several acute and chronic inflammatory, autoimmune, and malignant diseases. Although considered a genetically stable virus, recent sequence information obtained from a large number of viral isolates from around the world revealed that numerous viral variants exist. Whether these different strains differ in pathogenicity and immunogenicity and thereby contribute to the varying incidence rates of several EBV-associated diseases in different geographical regions is now studied intensively. The recent identification of amino acid sequence polymorphisms in a high percentage of all known virus-specific CD4+ and CD8+ T-cell epitopes, and of holes in the individual T-cell repertoire against epitopes derived from strain variants, may suggest that antiviral immunity is incompletely cross-protective against diverse EBV strains. These findings may have implications for immunological approaches seeking to prevent, monitor, or treat EBV-associated diseases.
Introduction
EBV is one of the most common viruses in humans and involved in the pathogenesis of several non-malignant and malignant diseases1,2. Primary infection with EBV usually occurs within the first three years of life by parent-to-child oral transmission. Following uptake, EBV is assumed to replicate in the oropharynx from where it colonizes the host by latently infecting B cells. The reservoir of latently infected cells can seed foci of virus replication at mucosal sites, and this sporadic reactivation allows the virus to persist for life in the infected host and to spread within the human population3. Carried by more than 90% of the worldwide population, EBV infection is usually asymptomatic and well controlled by T cells specific for epitopes derived from various latent and lytic cycle antigens of the virus4,5. However, a small percentage of virus carriers eventually develop EBV-associated disorders including malignant diseases that account for 1-2% of all human cancers1,2. Incidence rates of some EBV-associated diseases appear geographically clustered, such as nasopharyngeal carcinoma (NPC) and endemic Burkitt lymphoma that are highly prevalent in southern China and sub-Saharan Africa, respectively6,7. Together with the histological heterogeneity of tumor subtypes, these observations have led to the assumption that besides regional dietary, genetic, and environmental co-factors, EBV strains with distinct pathogenicity may exist in locally restricted areas and contribute to this geographic predilection2,8. Recent sequence analyses of a large number of whole virus genomes have identified numerous viral variants and provided new insights into virus transmission patterns and disease-associated genotypes9-12. To which extent sequence disparities among different EBV isolates impact on antiviral immunity, and whether immunotherapeutic and vaccine approaches that are based on a reference strain will be sufficiently cross-protective against strain variants, is yet to be discovered.
EBV strain variations
Based on their EBV nuclear antigen (EBNA) 2 and EBNA-3 gene sequences, EBV strains are grouped into type 1 or type 2 (formerly denoted as type A and B)1,2,6. Several groups have recently reported on the sequences of more than 150 viral isolates from different healthy virus carriers and tumor specimens. Approximately two thirds of these isolates were derived from tumor specimen (NPC, gastric cancer, Burkitt and Hodgkin lymphoma, etc.) and one third from non-malignant tissue (spontaneous LCL, PBMC, saliva). These studies identified complex variations in EBV strains within the type 1/type 2 classification system, and, when clustered geographically, a clear separation of Asian strains6,9-12. Moreover, a recent comparative analysis of whole genome sequences identified a number of polymorphisms that were shared by tumor-derived strains but absent in viral isolates from healthy study participants10. Together with the recent observation that the NPC-derived viral strain M818,13 shows enhanced epitheliotropism and lytic activity, these findings suggest that some sequence variations may result in altered pathogenicity. Furthermore, polymorphisms have been identified that result in amino acid exchanges in known T-cell epitopes9,10. Whether these sequence variations are by-products of random genetic drift, positively selected for reasons related to the function of the protein, or the results of an immune-driven selection process based on different MHC types that are prevailing in different geographical regions, is still unknown14.
EBV immunity and immunotherapy
EBV-specific T cells recognizing viral latent and lytic cycle antigens play a crucial role in antiviral immunity, including the control of EBV-driven lymphoproliferation4,5. Consequently, patients with primary or secondary T-cell deficiency and/or dysfunction are at risk of developing life-threatening EBV-associated lymphoproliferative disease. Reconstitution of EBV-specific immunity in these patients by the adoptive transfer of virus-specific T cells has been shown to be effective in preventing and treating such EBV-associated disorders15-19. Virus-specific T-cell lines for adoptive transfer were initially generated by repeated stimulation of peripheral blood T cells with autologous EBV-transformed lymphoblastoid B-cell lines (LCL) in vitro20. More recently, modified LCL were introduced as antigen presenting cells, e.g. LCL pulsed with synthetic peptide pools encompassing viral antigens, or transduced with vectors expressing viral proteins. Although safe and efficacious, LCL-based approaches take several weeks to yield a product suitable for clinical use. Therefore, new protocols for a faster and more efficient production of virus-specific T-cell lines have been developed. These are based on the selection of EBV-specific T cells with MHC-multimers, the immunomagnetic selection of IFNγ-secreting T cells after stimulation of peripheral blood mononuclear cells (PBMC) with viral peptide epitopes, or on using dendritic cells transfected with selected EBV genes to expand virus-specific T cells19-22. Although first clinical evidence indicates similar efficacy as traditionally manufactured virus-specific T-cell lines, such rapidly generated EBV-specific T-cell preparations are usually prepared using EBV epitope sequences from the widely used reference strain B95.84,23. Given the significant sequence diversity among EBV strains, these studies still left unanswered whether immunotherapeutic and vaccine approaches that rely on B95.8 epitopes are sufficiently cross-protective against other EBV strains.
Impact of EBV sequence variation on the T-cell response
Soon after variations in genome sequences were identified and EBV classified into type 1 and 2, differences in T-cell recognition of target cells infected with different viral strains were observed24. The notion that sequence diversity may affect immune recognition was substantiated when the first epitope recognized by EBV-specific CD8+ T cell was molecularly defined and shown to differ between the prototypic type 1 strain B95.8 and the prototypic type 2 strain AG876 in two amino acids25. With the identification of a large array of CD8+ T cell epitopes23, a complex picture of the antiviral T-cell response against different viral strains emerged. While several CD8+ T-cell epitopes were found to be conserved in different viral strains26, some were found to differ between virus types27, whereas others were reported to vary even within the same virus type28-33. Moreover, amino acid exchanges in CD8+ T-cell epitopes were shown to retain, reduce, or abolish T-cell recognition28-31,33,34. Collectively, these studies clearly demonstrated that sequence variations in EBV strains may reside within T-cell epitopes and potentially compromise the antiviral CD8+ T-cell response.
These studies, however, left unanswered, which proportion of the EBV-specific CD8+ T-cell epitopes is polymorphic, whether polymorphisms also occur in CD4+ T-cell epitopes and how they affect T-cell recognition, and whether the individual T-cell response encompasses specificities that recognize epitope variants. These questions were addressed in a recent study comparing 65 and 118 published epitopes recognized by EBV-specific CD4+ and CD8+ T cells, respectively, in the three distantly related viral strains B95.8, AG876, and M8135. When the type 1 strains B95.8 and M81 were compared to the type 2 strain AG876, more than 70% of all CD4+ and more than 40% of all CD8+ T-cell epitopes and/or their flanking regions were found to be polymorphic. Even between the two type 1 strains B95.8 and M81, more than 50% of all CD4+ T-cell epitopes and almost 40% of all CD8+ T-cell epitopes and/or their flanking regions proved to be polymorphic in that study35. As reported for CD8+ T cells, polymorphisms in CD4+ T-cell epitopes impaired T-cell recognition to various degrees35. Amino acid exchanges in epitopes or flanking regions may impact on T-cell responses in different ways. Examples of changes in MHC or TCR binding affinity33,34,36 and HLA restriction36 have been reported, but also different steps in antigen processing and presentation may be affected, e.g. cleavage by cytoplasmic or lysosomal proteases, transport of cytosolic peptides into the ER by TAP, and trimming of the peptides in the ER by ERAP37-39 (Figure 1).
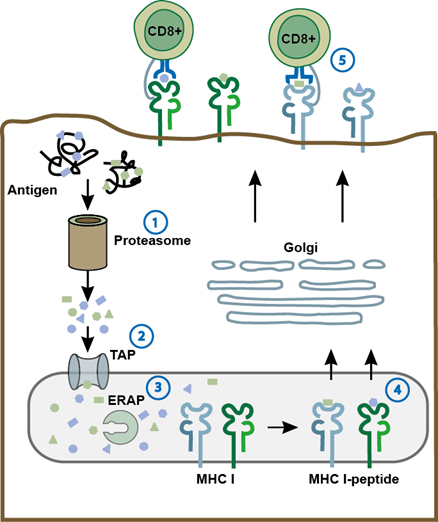
Figure 1: Amino acid exchanges in epitopes may affect antigen processing, presentation, and recognition by T cells. As shown for the MHC class I pathway, polymorphisms in epitopes may affect different steps in antigen presentation such as cleavage by the proteasome (1), transport by TAP (2), or trimming by ERAP (3). Also, MHC (4) and T-cell receptor binding affinity (5) may be affected. Similar mechanisms are thought to apply to MHC class II antigen processing and presentation. ERAP, ER aminopeptidase associated with antigen processing; TAP, transporter associated with antigen processing.
Moreover, the individual CD8+ T-cell response against polymorphic epitopes appears to vary between donors and to be focused on some epitope variants while others are ignored35. To which extent holes in the epitope-specific T-cell repertoire impair antiviral immunity and allow co-infection with strain variants is still unknown. Given the recent technical advances that facilitate full-length EBV genome sequencing from different tissues, first insights may be provided in the near future by comparing sequences of EBV genomes from individuals co-infected with more than one strain9-11,40,41. Similar experiments may also shed light on the role of the immune system on the time-dependent decrease in viral strain diversity as noted in IM patients40.
Potential clinical implications of viral strain heterogeneity
In patients with EBV-associated disorders, virus-specific T-cell responses are often monitored using overlapping peptide libraries covering mostly latent cycle proteins e.g. EBNA342,43. These libraries are based on B95.8 sequences and may incompletely measure virus-specific T-cell responses if the patient is infected with a different viral strain. Likewise, current approaches for the design of therapeutic EBV vaccines, and for the generation of virus-specific T-cell preparations for the treatment of EBV-associated disorders, are mostly based on latency proteins from B95.821,22,44-50. Since these proteins are highly polymorphic, clinical efficacy may be impaired should the pathogenic strain be divergent. For example, therapeutic EBV vaccines, which aim at increasing and sustaining the number of virus-specific T cells in persistently infected individuals, may fail to boost relevant T-cell responses in vaccinees infected with strain variants. Likewise, EBV-specific T-cell preparations for the treatment of EBV-associated disorders in stem cell or organ transplant recipients may show diminished clinical efficacy when T-cell donor and recipient are infected with different viral strains. Consequently, peptide-selected T-cell preparations for clinical use should be directed against a broad set of viral antigens to increase the likelihood of targeting shared epitopes. In addition, the incorporation of sequence information from all published virus isolates into epitope databases is expected to identify viral epitopes that are conserved in all strains or at least in subgroups, such as those prevalent in certain geographical regions. Alternatively, diagnostic methods for rapid virus typing in patients may be developed to facilitate the identification of customized sets of antigens for tailored therapy.
Conclusion
The identification of a growing number of EBV strains continues to add to a global view on EBV genome diversity and to a better understanding of EBV biology and the relationship between viral sequence variation and EBV-associated diseases. These studies have also identified substantial diversity in proteins targeted by the adaptive immune system. Incorporating information on sequence heterogeneity into immune monitoring assays, vaccine design, and immunotherapeutic approaches may ultimately improve clinical outcome of patients with EBV-associated disorders.
Acknowledgements
The authors declare that they have no conflict of interest.
Funding was received from the TUM Graduate Center and DZIF, the German Center for Infection Research (TTU 07.0804).
References
- Longnecker RM, Kieff E, Cohen JI. Epstein-Barr virus. in Fields Virology (eds. Knipe, D.M. & Howley, P.M.) 1898-1959 (Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, 2013).
- Rickinson, AB, Kieff E. Epstein-Barr Virus. in Fields Virology (eds. Knipe, D.M. & Howley, P.M.) 2575-2627 (Lippincott Williams & Wilkins, Philadelphia, 2007).
- Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. The New England journal of medicine. 2004; 350: 1328-1337.
- Taylor GS, Long HM, Brooks JM, et al. The immunology of Epstein-Barr virus-induced disease. Annual review of immunology. 2015; 33: 787-821.
- Long HM, Taylor GS, Rickinson AB. Immune defence against EBV and EBV-associated disease. Current opinion in immunology. 2011; 23: 258-264.
- Farrell PJ. Epstein-Barr Virus Strain Variation. Current topics in microbiology and immunology. 2015; 390: 45-69.
- Rickinson AB. Co-infections, inflammation and oncogenesis: future directions for EBV research. Seminars in cancer biology. 2014; 26: 99-115.
- Feederle R, Klinke O, Kutikhin A, et al. Epstein-Barr Virus: From the Detection of Sequence Polymorphisms to the Recognition of Viral Types. Current topics in microbiology and immunology. 2015; 390: 119-148.
- Palser AL, Grayson NE, White RE, et al. Genome diversity of Epste in-Barr virus from multiple tumor types and normal infection. Journal of virology. 2015; 89: 5222-5237.
- Zhou L, Chen JN, Qiu XM, et al. Comparative analysis of 22 Epstein-Barr virus genomes from diseased and healthy individuals. The Journal of general virology. 2017; 98: 96-107.
- Kwok H, Wu CW, Palser AL, et al. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. Journal of virology. 2014; 88: 10662-10672.
- Yao Y, Xu M, Liang L, et al. Genome-wide analysis of Epstein-Barr virus identifies variants and genes associated with gastric carcinoma and population structure. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017; 39: 1010428317714195.
- Tsai MH, Raykova A, Klinke O, et al. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell reports. 2013; 5: 458-470.
- Tzellos S, Farrell PJ. Epstein-barr virus sequence variation-biology and disease. Pathogens. 2012; 1: 156-174.
- Heslop HE, Rooney CM. Adoptive cellular immunotherapy for EBV lymphoproliferative disease. Immunol Rev. 1997; 157: 217-222.
- Gottschalk S, Rooney CM. Adoptive T-Cell Immunotherapy. Current topics in microbiology and immunology. 2015; 391: 427-454.
- Merlo A, Turrini R, Dolcetti R, et al. Immunotherapy for EBV-associated malignancies. International journal of hematology. 2011; 93: 281-293.
- Bollard CM, Gottschalk S, Torrano V, et al. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014; 32: 798-808.
- McLaughlin LP, Bollard CM, Keller MD. Adoptive T Cell Therapy for Epstein-Barr Virus Complications in Patients With Primary Immunodeficiency Disorders. Frontiers in immunology. 2018; 9: 556.
- Rooney CM, Smith CA, Ng CY, et al. Infusion of cytotoxic T cells for the prevention and treatment of Epstein-Barr virus-induced lymphoma in allogeneic transplant recipients. Blood. 1998; 92: 1549-1555.
- Moosmann A, Bigalke I, Tischer J, et al. Effective and long-term control of EBV PTLD after transfer of peptide-selected T cells. Blood. 2010; 115: 2960-2970.
- Bollard CM. Improving T-cell therapy for epstein-barr virus lymphoproliferative disorders. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013; 31: 5-7.
- Hislop AD, Taylor GS, Sauce D, et al. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annual review of immunology. 2007; 25: 587-617.
- Moss DJ, Misko IS, Burrows SR, et al. Cytotoxic T-cell clones discriminate between A- and B-type Epstein-Barr virus transformants. Nature. 1988; 331: 719-721.
- Burrows SR, Sculley TB, Misko IS, et al. An Epstein-Barr virus-specific cytotoxic T cell epitope in EBV nuclear antigen 3 (EBNA 3). The Journal of experimental medicine. 1990; 171: 345-349.
- Burrows SR, Misko IS, Sculley TB, et al. An Epstein-Barr virus-specific cytotoxic T-cell epitope present on A- and B-type transformants. Journal of virology. 1990; 64: 3974-3976.
- Schmidt C, Burrows SR, Sculley TB, et al. Nonresponsiveness to an immunodominant Epstein-Barr virus-encoded cytotoxic T-lymphocyte epitope in nuclear antigen 3A: implications for vaccine strategies. Proceedings of the National Academy of Sciences of the United States of America. 1991; 88: 9478-9482.
- Apolloni A, Moss D, Stumm R, et al. Sequence variation of cytotoxic T cell epitopes in different isolates of Epstein-Barr virus. European journal of immunology. 1992; 22: 183-189.
- Lee SP, Thomas WA, Murray RJ, et al. HLA A2.1-restricted cytotoxic T cells recognizing a range of Epstein-Barr virus isolates through a defined epitope in latent membrane protein LMP2. Journal of virology. 1993; 67: 7428-7435.
- Midgley RS, Bell AI, Yao QY, et al. HLA-A11-restricted epitope polymorphism among Epstein-Barr virus strains in the highly HLA-A11-positive Chinese population: incidence and immunogenicity of variant epitope sequences. Journal of virology. 2003; 77: 11507-11516.
- Gras S, Chen Z, Miles JJ, et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. The Journal of experimental medicine. 2010; 207: 1555-1567.
- Bell MJ, Brennan R, Miles JJ, et al. Widespread sequence variation in Epstein-Barr virus nuclear antigen 1 influences the antiviral T cell response. The Journal of infectious diseases. 2008; 197: 1594-1597.
- Brooks JM, Croom-Carter DS, Leese AM, et al. Cytotoxic T-lymphocyte responses to a polymorphic Epstein-Barr virus epitope identify healthy carriers with coresident viral strains. Journal of virology. 2000; 74: 1801-1809.
- Burrows JM, Burrows SR, Poulsen LM, et al. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. Journal of virology. 1996; 70: 2490-2496.
- Cirac A, Stützle S, Dieckmeyer M, et al. Epstein-Barr virus strain heterogeneity impairs human T-cell immunity. Cancer immunology, immunotherapy : CII. 2018; 67: 663-674.
- Liu YC, Chen Z, Neller MA, et al. A molecular basis for the interplay between T cells, viral mutants, and human leukocyte antigen micropolymorphism. The Journal of biological chemistry. 2014; 289: 16688-16698.
- Ossendorp F, Eggers M, Neisig A, et al. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996; 5: 115-124.
- Rossjohn J, Gras S, Miles JJ, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annual review of immunology. 2015; 33: 169-200.
- Neefjes J, Ovaa H. A peptide's perspective on antigen presentation to the immune system. Nature chemical biology. 2013; 9: 769-775.
- Weiss ER, Lamers SL, Henderson JL, et al. Early Epstein-Barr Virus Genomic Diversity and Convergence toward the B95.8 Genome in Primary Infection. Journal of virology. 2018; 92.
- Correia S, Palser A, Elgueta Karstegl C, et al. Natural Variation of Epstein-Barr Virus Genes, Proteins, and Primary MicroRNA. Journal of virology. 2017; 91.
- Lam JKP, Hui KF, Ning RJ, et al. Emergence of CD4+ and CD8+ Polyfunctional T Cell Responses Against Immunodominant Lytic and Latent EBV Antigens in Children With Primary EBV Infection. Frontiers in microbiology. 2018; 9: 416.
- Korber N, Behrends U, Hapfelmeier A, et al. Validation of an IFNgamma/IL2 FluoroSpot assay for clinical trial monitoring. Journal of translational medicine. 2016; 14: 175.
- Taylor GS, Steven NM. Therapeutic vaccination strategies to treat nasopharyngeal carcinoma. Chinese clinical oncology. 2016; 5: 23.
- Smith C, Khanna R. The Development of Prophylactic and Therapeutic EBV Vaccines. Current topics in microbiology and immunology. 2015; 391: 455-473.
- Cohen JI. Epstein-barr virus vaccines. Clinical & translational immunology. 2015; 4: e32.
- Bollard CM, Heslop HE. T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood. 2016; 127: 3331-3340.
- Icheva V, Kayser S, Wolff D, et al. Adoptive transfer of epstein-barr virus (EBV) nuclear antigen 1-specific t cells as treatment for EBV reactivation and lymphoproliferative disorders after allogeneic stem-cell transplantation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013; 31: 39-48.
- Haque T, Wilkie GM, Jones MM, et al. Allogeneic cytotoxic T-cell therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood. 2007; 110: 1123-1131.
- O'Reilly RJ, Prockop S, Hasan AN, et al. Virus-specific T-cell banks for 'off the shelf' adoptive therapy of refractory infections. Bone marrow transplantation. 2016; 51: 1163-1172.
