Importance of Interkingdom Interactions Among Oral Microbiome Towards Caries Development – A Review
Kalpana Balakrishnan1, 4, Divya Sivanesan1, Gaanappriya Mohan4, Sachin S Gunthe2,3, Rama S Verma1*
1Department of Biotechnology, Indian Institute of Technology Madras, Chennai
2Department of Civil Engineering, Indian Institute of Technology Madras, Chennai
3Laboratory of Atmospheric and Climate Sciences, Institute of Technology Madras, Chennai
4Department of Biotechnology, K. S. Rangasamy College of Technology, Namakkal Tamil Nadu, India
Abstract
The human microbiome plays a crucial role in health and disease conditions. These microbiomes constitute a structured, coordinated microbial network throughout the human body. The oral cavity harbors one of the extensively diverse bacteria in the human system. Although many studies emphasize bacteriome and its interaction with the host system, very little attention is given to candidate phyla radiation (CPR), fungal components, and its interkingdom interaction in the oral microecology even with advanced techniques. The interkingdom interactions among caries causing microbes trigger the pathogenesis of bacterial diseases and cause ecological shifts and affect the host system. Studying the complex relations among the diverse oral microbiome and its host, especially CPR phyla and fungi, would give a holistic view of the caries etiology. This review provides evidence on the interkingdom interaction that establishes a complex community that could help predict future oral and systemic diseases.
Introduction
Metagenomic analysis through high-throughput sequencing technologies has revolutionized the study of the human microbiome, especially the oral system. To date, 700 species or phylotypes of microbes were reported to harbor the oral microbiome1,2. Site-specific microbial profiling has demonstrated that microecological nature at different sites like the teeth, tongue, buccal mucosa, hard and soft palate inhabits distinct microbial communities. Colonization of the microbes is mediated by surface nature for microbial adherence, oxygen availability, host defense, pH, temperature, nature of food intake and salivary flow3–5. The oral microbiome exhibits a complex interdependent taxonomic community that plays a crucial role in an individual's health. The communication among microbes, such as direct cell to cell interaction via chemical signals and metabolic cooperation, leads to biofilm formation6. Shift in the microecological balance due to microbiome dysbiosis because of high sugar-rich dietary intake enhances the exopolysaccharide production via quorum sensing signals mediating a strong architectural biofilm formation that leads to caries7,8. Analysis of the caries microbiome revealed that 70% of the diversity belongs to the high abundant core microbiome. The less abundant species contribute to greater diversity that was found to be site-specific, termed as 'rare-biosphere'9,10. A decade of studies on the caries microbiome focused mainly on bacteriome and recently on mycobiome; however, there is still no emphasis on the interkingdom interactions.
The cross-kingdom synergies are involved in the pathogenesis of both mucosal and dental diseases11. Complex physical and chemical interactions (including cross-feeding and metabolites exchange), as well as environmental and host factors, govern the development of pathogenic bacterial–fungal biofilms12. Such formed biofilm provides spatial organization for other bacteria, virulence, and drug protection or resistance that results in recurrence13,14. It is of rising interest in current caries research that how this interkingdom interaction under a specific niche shifts the healthy symbiosis to dysbiosis and how it helps to persist any treatment methods leading to disease. The oral cavity's microbiome is an essential source in many oral and systemic diseases15,16. A study on the recurrence of disease in denture stomatitis, lungs of cystic fibrosis and immunocompromised patients, recurrent bowel disease, infections of burn wounds reports the shift in microecology due to interkingdom interactions17,18. Additionally, not much is known about the spectrum of the bacteriome ecological relationships with mycobiome and CRP phyla and its host. This review summarizes recent work to elucidate the oral microbiome's ecological shifts due to interkingdom interactions, focusing on bacteria, fungi and CRP phyla.
Human Oral Microbiome
Oral microbiota, oral microflora, or oral microbiome is termed to denote microorganisms' conglomerate in the human oral cavity. Microbiota refers to the interacting microbes in any given environmental conditions, while microflora used to differentiate a sub-group of organisms from the whole microbiota. In contrast, the term microbiome refers to the microorganism, its genetic material and its contribution to the human's health. The word microbiome was coined by Joshua Lederberg to elucidate the ecological community of mutualistic, symbiotic, and pathogenic microorganisms in our body. The term has been integrated by the Human Microbiome Project and investigators to comprehend human health and disease19.
The oral cavity is one of the profoundly occupied colonies in the human body. The current human oral microbiome encompasses heterogeneous microbial communities composed of few hundreds of bacterial species - belonging to the Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria3 and few fungal species, principally Candida, Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus and Fusarium18. The oral microbiome is still investigated upon for its potential effects on human lifestyle, disease-causing / prevention and on oral hygiene20,21. Enduring host-microbiome relationships are established on adaptive strategies within an ecological niche. So, explorations are intended at establishing the mechanisms of interactions between the microbiome and humans22. The scientific community is beginning to recognize that the acquirement process of the wide-ranging species assimilating the ‘normal’ and beneficial microbiome is an essential process for oral health, but it is still not completely obvious how and when it happens and how it is regulated23.
Composition of the Oral Microbiome
Ever since the Human Microbiome Project (2007) was launched, a flux of research paved the way to the exploration, identification, and thorough analysis of numerous microorganisms of which the commonest was explicatory of the role of the microbiome in disease pathology. Several works and research have intended to ascertain the dysbiosis connected with diseases rather than focusing on the microbiome states and characters and activities related to human health. Because ‘healthy’ microbiomes are for better health, it is certainly indispensable for us to identify, group, characterize, and define the microbiomes of health and harness their potential in commensals' therapeutics.
- Viruses: A wide range of pathogenic viruses could be located in the oral cavity. Contagious pathogens that are transmitted via the salivary juices are located in the oral cavity of infected persons. Bloodborne viruses and respiratory infection causing viruses are also harbored in the oral cavity. An oral virome study by Pride et al.24 portrayed the homology of the identified order to be bacteriophages which is not a startling result due to the density of bacterial population in the oral cavity.
- Protozoa: As part of the normal microbiome, protozoans like Entamoeba gingivalis and Trichomonas tenax are recognized in healthy individuals. These organisms breed in poor oral hygienic conditions. Currently categorized as harmless saprophytes, these organisms can change into potential pathogens leading to diseased circumstances25.
- Fungi: The basal oral microbiome had 74 culturable and 11 non-culturable fungal genera in fit humans18. Candida was the most popular species, and moreover, were a cluster of fungal species reported, including cerevisiae, Penicillium, Geotrichum, Aspergillus, Scopulariopsis, Hemispora, Hormodendrum. Fungal species distribution is diverse phenomenally in the oral cavity between healthy individuals. Aspergillus, Fusarium, and Cryptococcus isolates were witnessed in the oral cavity of healthy humans. Such studies show that the other microbial species could have an internal ecosystem that limits the fungal genera's pathogenicity in healthy persons in the oral microbiome and the immune systems of the healthy persons18.
- Archaea: Archaea are well-noticed constituents of the human microbiome. However, these microbial species are largely under-represented when compared to the larger diversity of bacterial taxa. Methanobrevibacter oralis thrives in various niches within the oral cavity26.
- Bacteria: Due to limitations in the methods to elucidate the microbiome, fewer bacteria have been identified and characterized from the oral cavity of a healthy individual. The oral bacterial community of healthy humans is controlled by Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes, and Fusobacteria, which account for 96% of species detected to date3. Species wise, Streptococcus3,27, Neisseria spp, Bacillus spp., Rothia spp during diseased conditions4 Granulicatella species during both diseased and healthy environments27,28 are mainly seen.
Adult and Childhood Caries
Across the globe, dental caries are considered a chronic condition in people of all ages. Some caries are treated at early stages. However, the unidentified, untreated or ignored caries lead to pain and infections that spread intensively, culminating with intensive treatments and cure. The occurrence of carbohydrates and biofilms in the oral cavity and teeth supports the metabolism of microorganisms. The metabolic process marks the discharge of acidic by-products, which dissolve the hydroxyapatite constituents of the enamel, dentin and cementum. This demineralization points to cavities on the tooth surface. As days proceed, it becomes hard to eliminate the sturdy biofilms on the cavitated surfaces, thereby augmenting the multiplication of microbes in the environment. Be it in adults or children, the dentition process pints to rapid progression of caries and hence restorative dentistry is always recommended29. Nevertheless, in the early stages of dental caries, our body has a repair mechanism for the demineralization in the cavities, and this process is termed as remineralization. Remineralization occurs when the minerals in the saliva disperse to the porous surfaces, and the natural restoration begins30. Whether a lesion will progress, continue to remain the same, or reverse is decided by the balance between protective factors and pathological factors, which is termed as the ’caries balance’31.
Studies expose that neonatal factors increase the risk of caries acquirement at an early age due to the transmission of S.mutans32–36. It is hypothesized and studied that infants delivered by caesarean section obtain the infection than the vaginally delivered infants37. Systemic reviews propose that dental caries is affected by several factors, including good oral hygiene, socioeconomic status, poverty, deprivation, non-carcinogenic diets, and ethnicity.
Many effective strategies are existing to thwart and treat caries in adults and children. ’Active surveillance’ is a term that is keenly used to elucidate the observation of caries in children and adults. The dentists propose disease management plans based on the patient’s individual factors. Some of the active surveillance strategies include brushing twice daily with toothpaste containing fluoride – which results in enhancing the remineralization process, making alterations in the intake of sugars, flossing actively for adults, etc., are definitely noted strategies in caries management and prevention 38.
Authenticating the effectiveness of fluoride in preventing/managing dental caries among adults is important. On a population basis, caries fetches a more significant health problem among adults, especially older adults, since they are more likely to retain their natural teeth than in preceding generations. Reviews suggest that behaviour modification is effective in managing caries; however, bringing in a behavioural change is extremely challenging. Efforts for an engagement at the community, family, and individual level, based on delivery of information and skill training, have had diverse results to date39. A remarkable finding by Griffin et al.40 was the several modes of fluoride delivery among adults and their similarity to findings in children. One probable reason for the lack of preventive programs for adults may be the lack of evidence on their effectiveness for this population41.
Techniques Used in the Identification of Oral Microbiome
A range of traditional strategies was utilized to study the oral microbiome composition, comprising microscopy, cultural analysis, enzymatic assays, and immunoassays. Preceding researches have characterized the oral microbiome of historic European cultures42. Outcomes have proven that changes in nutritional conduct related to specific time intervals affect the human oral microbiome's structure and function43. Pre-Columbian cultures also had a substantial impact on cutting-edge western societies, and characterization in their oral microbiome may furthermore provide insights into the ancestral state of the human oral microbiome for the duration of a part of human history44. Modern molecular taxonomic approaches, particularly the ability to rapidly obtain large numbers of 16S ribosomal DNA sequences, have shown the path for exhaustive oral microflora surveys.
Several oral bacteria are fastidious and slow-growing, calling for very complex growth media and incubation conditions. Few are strict anaerobes, and henceforth in vivo cultural analysis of samples is hard and only permits for the processing of trifling sample numbers. Discerning bacteriological media have confirmed only a few interest species valuable for studies, while the others remain undetected. Figure 1 expounds on the identification methods of microbes over a period of time. Over the past 15 years, through numerous oral microbiologists' collaborative endeavours, 68% of oral bacterial species inside the mouth have been cultured. This differentiates with a long way to reduce the probability of microorganisms cultured from the skin or gut microflora45. When a bacterium is cultured, it can be characterized completely, used in in vivo and in vitro experiments, handled by culling out or swapping genes, and officially named. The use of ribosomal RNA gene sequences as phylogenetic markers revolutionized the investigation of molecular evolution, phylogeny, and ecology in all living organisms. The oral microbiome has been remarkably characterized, each via cultivation as well as cultureâindependent methods. Culture-independent approaches, together with the 16S rRNA geneâprimarily based molecular strategies, have changed mainly cultivation research because molecular strategies can display the identities of currently uncultivated microorganisms. Subsequently, our appreciation of microbial variety has received an impetus from SSU (Small Sub Unit) rRNA gene analyses based on the 16S ribosomal RNA gene of bacteria and archaea and the 18S ribosomal RNA gene of fungi, especially the ITS (Internal Transcribed Spacer) loci, describing a phylogenetic framework for the type and evaluation of microbial range in any given surroundings without the requirement for isolation and cultivation46.
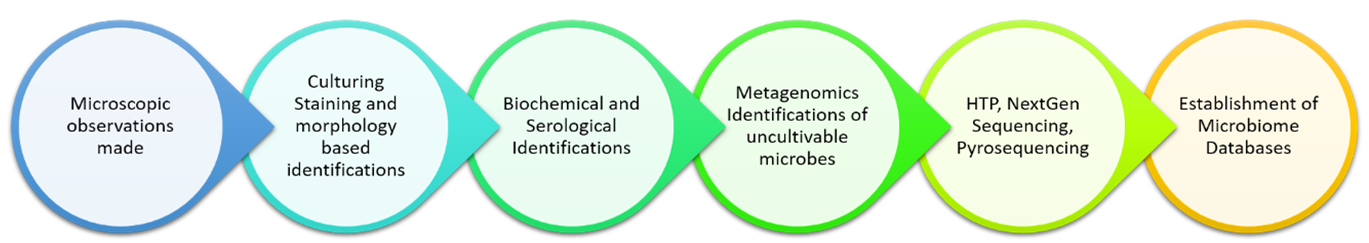
Figure 1: Schematic Illustration on Technological Improvement in the process of microbial identification
Contemporary studies often depend on 16S rRNA gene profiling via PCR amplification with general SSU rRNA gene primer sets to provide an overview of a microbial community or microbiome's taxonomic composition. This method has changed our understanding of global microbial diversity and exponentially increased the number of 'uncultivated' representatives of the universal phylogenetic tree47,48.
The traditional 16S rRNA gene sequencing method had its drawbacks: costly, time-consuming, and highly laborious. The start of Next Generation Sequencing methods such as 454 pyrosequencing and Illumina MiSeq have facilitated an increase in the sample throughput, with up to 27 million sequences being generated in a single run49,50. That ease and reasonably cheap NGS prompted an enormous information era leading to vast research publications detailing genetic sequences and interpretations. Important consideration should be taken while claiming NGS investigations to avoid the samples' contamination through the extraction kits, lab reagents, and test gathering tools, which affects the studies' consequence. Other day-to-day methods comprise metagenomics and metatranscriptomics. Metagenomics focus to explicate the whole microbial community's genetic compositions in the samples; metatranscriptomics let the researchers study the microbial community's vigorously transcribed genes without independently culturing the microorganisms. Researchers have studied the density and distribution of antibiotic protein in human oral and stool microbiome datasets using a specially constructed profile Hidden Markov Model (HMM)51. Such methods are, to date, still impeded by technical challenges but offer ample opportunities to further our knowledge of the collective genome of the oral microbiome and its future metabolic actions.
Oral bacteria typically live as part of a multispecies community in thickly populated biofilms. There are many gradients of nutrients within the biofilm, signaling molecules and gases occurring due to the diffusion patterns of these substances and neighboring bacteria's metabolic activity. For individual cells and groups of cells, they can vary markedly52,53. Culture-independent methods have thrown light on the microbiome's diversity, but to elucidate the microbes' characters and constraints, they are grown in culture. Currently, new approaches are being resorted to growing the uncultivable microbes in the lab. Siderophores, scaffolds, and other biometrics focus to be applied as a substratum for developing these novel strains54. Few isolates depend on 'helper strains' for thriving culture, implying their dependency on quorum-sensing and nutritional and/or signaling interactions with other bacteria within the biofilm community that they naturally inhabit. The application of such novel culturing methods reinforced with emerging molecular, bioinformatic techniques and increased computational power will intensify the oral microbiome's understanding and give information to devise interventional approaches to target the diseases.
Ecology of Oral Microbiome
The mouth covers various surface types, including un-keratinized and keratinized gingival epithelium and hard tissues as teeth. Gingival crevicular fluid and saliva, along with several other fluids, also settle in the mouth from time to time. The oral cavity is open to materials from the surroundings, including foodstuffs and debris sampled from items placed within the mouth (fingers, utensils, etc.). So, the prospects of harboring several microorganisms are very high, leading to the formation of an oral biofilm, which has been studied extensively. The attachment of the microbe initiates the formation of oral biofilm to the host surface. After the attachment, the microbes divide and secrete polymers to form a scaffold to dock multiple microorganisms55.
The oral microflora encompasses ecological surface niches or the earlier defined biofilms that evolve in due course of human life: first of all, as microbial populations adherent to mucosal surfaces exceeded on from the parental flora to enamel-adherent populations following the outbreak of the dentitions, and with changes in each supra- and subgingival niche (dental plaque/biofilm)56–58. In diseased conditions, there may be a shift inside the equilibrium far from the dynamic, synergistic interaction of these healthy oral microbial populations in the direction of a narrower diversity of healthy populations in opposed interaction with pathologic populations with variable inflammatory host immune responses58. The shape and characteristics of the oral microflora (and related microbiome) have been examined in many oral diseases resulting from bacteria, fungi, and viruses (e.g., Periodontal illnesses) and the systemic diseases associated with continual infections (e.g., Diabetes mellitus, cardiovascular disorder, and most cancers)59–61.
Interestingly, the interaction of microbiome in the oral cavity with that of gastrointestinal tract has been reported in many systemic diseases such as liver cirrhosis62. The striking part of the finding was that the oral microbiome invades and colonizes the gut during disease condition, but no sign of the gut microbiome invading the oral cavity was seen. Many articles have shown that this transmission plays a vital role in colorectal and gastrointestinal cancer risk19,45,62. The probability of cancer is more in people with the dental disease as oral bacteria activates alcohol and other carcinogens63. Furthermore, a detailed study on how presence of oral microbiota in gut can be used as a biomarker for many inflammatory bowel diseases like Crohn’s disease has been published recently64. This can take us one step further in having better prognosis for gastrointestinal diseases.
The oral ecosystem is an unstable microbial community that frequently interacts and influences the surrounding factors. The dynamics between the oral environment and the oral microbial community has to be essentially studied practically to understand the colonizing microbes at various sites, the metabolism of the microbes, and the physicochemical factors that impact the harboring of oral microbes in the oral cavity65. Factors that influence microbial composition include genetics, host defenses, microbial interactions, receptors used for attachment, temperature, pH of the mouth, oxidation-reduction potential, availability of nutrients and water, acidogenicity, and salivary flow4.
The key to oral health is an ecologically stable and varied microbiome that practices commensalism within itself and mutualism with its host. In a healthy oral cavity, an ecological balance exists between the host and the numerous indigenous microorganisms66. This relationship allows them to flourish, maintain biodiversity within the oral cavity and keep their host healthy. The interactions can also repress functions of the member species to modulate population growth, biofilm structure, community changes, and spatial organization67. The shifts in relationships, proportion and virulence properties of microbes appear to affect one another, and consequently, it is not sure which ecological shift happened first. It is also uncertain what precisely activates the initial ecological shift and, in turn, catalyzes the entire cycle68.
The vital factors for starting an ecological shift are poor oral hygiene, compromised immune system and genetics. Genetic factors also play a central role in convincing the immune system formation, which could either enhance beneficiary microbes’ survival or create a physical and physiological environment that aid pathogenic microbes survival69. For instance, in the case of humans suffering from hyper-IgE inflammatory syndrome who develop oropharyngeal candidiasis, streptococcus oralis and S. mutans are the most70. The oral cavity's microbiome is an indispensable source in many oral and systemic diseases. Poor oral hygiene is a reason for the accumulation of bacteria within biofilms. When these bacteria do not detach, it results in the formation of plague, and pathogenicity increases, causing dental diseases44.
Ecological Shifts in the Oral Microbiome
The key to oral health is an ecologically balanced and diverse microbiome that practices commensalism within itself and mutualism with its host. In a healthy oral cavity, an ecological balance exists between the host and the numerous indigenous microorganisms71. This relationship allows them to flourish, maintain biodiversity within the oral cavity and keep their host healthy. The interactions can also repress functions of the member species to modulate population growth, biofilm structure, community changes, and spatial organization8,72 Figure 2. The shifts in relationships, proportion and virulence properties of microbes seem to affect one another, and therefore, it is not sure which ecological shift occurred first. It is also unclear what exactly triggers the initial ecological shift and, in turn, catalyzes the entire cycle73.
The crucial factors responsible for initiating an ecological shift are poor oral hygiene, compromised immune system, and genetics. Poor oral hygiene is a cause for the accumulation of bacteria within the biofilms and when these bacteria do not detach, it leads to the formation of plaque, and pathogenicity increases, causing dental diseases74. Genetic factors also play a significant role in inducing the immune system formation, which could either enhance beneficiary microbes’ survival or create a physical and physiological environment that aid pathogenic microbes survival75. For instance, in case of humans suffering from hyper-IgE inflammatory syndrome who develop oropharyngeal candidiasis, Streptococcus oralis and S. mutans are the most abundant during active fungal infections76. Candida albicans was found to be dominant in terms of abundance and ubiquity in the childhood oral microbiome. Interestingly they also found 17 fungal species that were significantly abundant in healthy dentitions, whereas very few were enriched in dental caries in children, suggesting an associated shift in dental biofilm, which makes them less favorable for the survival of many fungi77. Candida, along with increased aciduric bacteria, may increase the risk of oropharyngeal candidiasis78. Among the phyla studied, Firmicutes and Actinobacteria were the major taxa found in caries affected children79.
Interaction among the microbes is mediated by quorum sensing molecules (QSM) to regulate a wide range of behavior patterns among them. The in vitro studies reveal that more studies on quorum sensing will help us understand the basic machinery of cell-cell signaling in microbial communities. The free-floating microbiomes end up having high levels of metabolic by-products and other secondary metabolites, which also contain quorum sensing molecules20. These QSM function through the secretion and detection of autoinducer molecules that accumulate based on cell density. When the AI molecules increase in number, the bacterial receptors are activated, leading to signal transduction cascade resulting from which specific genetic set up is switched on in the plague films or the floating bio-films80. Quorum sensing is widely employed by a variety of gram-positive and gram-negative bacterial species to coordinate communal behavior67,36.
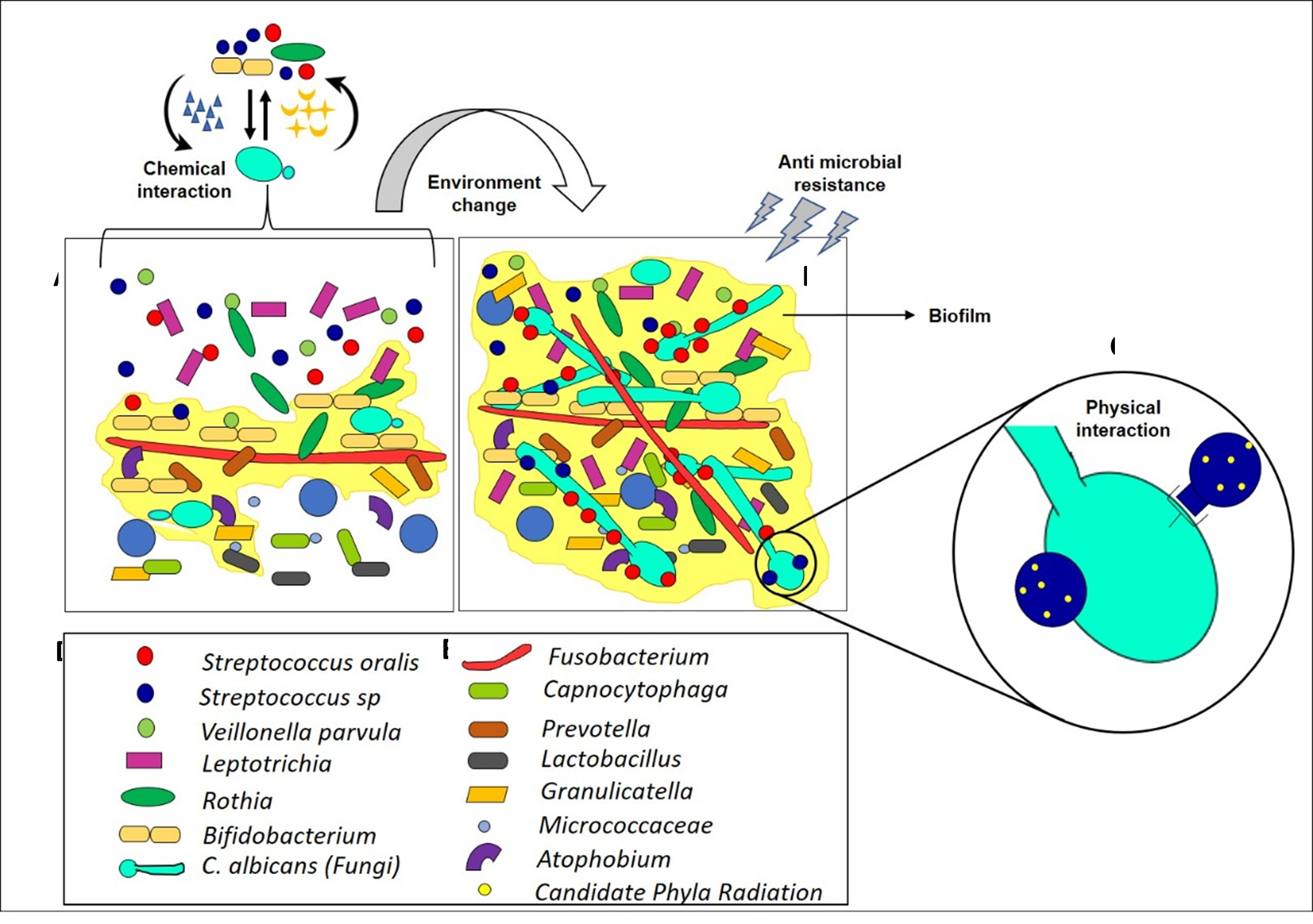
Figure 2: Schematic Illustration on Interkingdom Interaction of Caries free and Caries Oral Microbiome. The figure depicts the change in abundance of oral microbiota from caries-free (A) to caries (B). The metabolites produced by the bacteria and C.albicans mutually enhance its growth and invites more acidophilic and aciduric microbiota to interact with each other, which creates an acid- based niche resulting in caries formation. A: Interaction of microbes in the healthy oral cavity that gives very low biofilm formation. B) Enhanced biofilm formation (yellow colour) due to complex interaction of microbes in the presence of C.albicans. C) Representation of interkingdom interaction among C.albicans (green), S.oralis (Blue), Candidate Phyla Radiation (Yellow). D) List of early colonizers E) List of middle and late colonizers.
Bacteria Fungal Interaction
Oral bacteria and fungi have a wide array of interactions ranging from synergistic to antagonistic, impacting host immune response and health both positively and negatively9,81. The culture-independent analysis reported more than 75 diverse fungal species in a healthy oral cavity than the disease. Candida, Cladosporium, Aureobasidium, and Aspergillus constitute the most abundant, found in 25–75% of subjects with significant variations between individuals2,82. In bacterial-fungal interaction, fungi form a structural skeleton to a diverse spectrum of bacteria in multispecies biofilm with its property to generate filamentous hyphae and its larger cell size despite its low abundance12,83. Interaction between C. albicans and Streptococcus mutans or Streptococcus oralis to aggravate the severity of dental caries has been studied extensively. Although some researchers suggest that S. mutans and C. albicans relationship is mediated by glucans produced by functional glucosyl-transferases bound to C. albicans surface, it is still unclear whether the bacteria influences virulence or whether the fungus induces changes in bacterial diversity. Moreover, the interaction of C.albicans and Fusobacterium nucelatum helps to evade the host immune system emphasizing its increased importance in polymicrobial diseases76. Biofilm formed by the mutualistic effect of these cross-kingdom interactions increases microbial carriage and infectivity as well as enhances an extraordinary antimicrobial resistance.
Bacterial Viral Interaction
Little is known about the part of the components of viruses as affiliates of the human microbiome. We examined the composition of human oral viral communities in a cluster of relatively periodontally healthy subjects or significant periodontitis to determine whether health status may be associated with differences in viruses84. The viruses inhabiting dental plaque were pointedly different based on the oral health status, while those present in saliva were not. Dental plaque viruses in periodontitis were foreseen to be suggestively more likely to kill their bacterial hosts than those found in healthy mouths. Because oral diseases such as periodontitis have been exposed to have transformed bacterial communities, we believe that viruses and their role as drivers of ecosystem diversity are significant contributors to the human oral microbiome in health and disease states. In addition to investigating presence and abundance, community sequencing data helps identify potentially interacting species through co-occurrence or co-exclusion data. By analyzing the oral mycobiome of HIV patients, an antagonistic relationship between two fungal species based on their anti-correlation were ascertained85.This type of data can offer hypotheses for further physiological testing of potential interactions between organisms. By concurrently sequencing the bacterial, fungal, and viral communities, novel inter-kingdom interactions can be identified. Bacteriophages are significant drivers of bacterial diversity in various ecosystems86,87 and most of the viruses identified in saliva and dental plaque were phage. Lysogenic oral viruses live in dynamic equilibrium with their cellular hosts and, as a consequence, are highly persistent members of the human oral microbiome. In the development of periodontal disease, the surfaces of the gums and bones pull away from the teeth, forming pockets that are generally inhabited by different bacteria. While the profound differences in the subgingival virobiota may merely reflect changes in the bacterial biota that colonize these exposed pockets, the differences in the subgingival plaque virome in subjects with periodontal disease may also have other biological implications. The lytic phage in the subgingival crevice probably helps shape the local microbiota and contribute to the local microbial community structure and change local biodiversity88.
Bacteria CPR Interaction
High-throughput sequencing technology frequently detects the three human-associated CPR phyla, Gracilibacteria (GN02), Absconditabacteria (SR1) and Saccharibacteria (TM7), in multiple body sites, including the oral cavity8,16. Among the three, TM7 is prevalent in the oral cavity and other mucosal diseases like vaginosis, halitosis, inflammatory bowel disease, and periodontitis89. TM7 employs an epiparasitic interaction with the host bacteria in-search of the essential amino acid88(Fig:2). This epiparasitic interspecies interaction would have a notable impact on microecology due to direct reciprocal effects on physiology and pathogenicity, thereby indirectly influencing the overall structural and functional complexity of oral microbiota and disease56. Further investigations using genomics, transcriptomics and metabolomics interactions will give a better insight into this intriguing relationship and its clinical impact on CPR organisms.
Conclusion
This review precises the progression of metagenomics study in understanding the complex microbial community inhabiting the oral cavity, how the interspecies and interkingdom interactions lead to ecological shift causing diseases remain understudied. Several recent reports started to explore the interspecies interactions, their co-occurrence and mutual exclusion of several bacteria in the oral cavity, while Candida albicans and TM7 were the only phyla explored for fungi and CPR species, respectively. A comprehensive understanding of other fungi and CPR phyla and their interaction with bacteria along with the host system will shed light on the etiology of microbes within the oral cavity. In summary, this review provides a holistic view of how microbial persistence leads to the recurrence of oral disease.
Acknowledgement
We extend our heart filled thanks to the Management and Principal of K S Rangasamy College of Technology for providing infrastructure to carry out this work. The corresponding author acknowledges DST-FIST, DBT- STAR scheme for providing infrastructure facilities. We thank all the previous researchers and their findings which has led us to this output.
Conflict of Interests
The authors declare no competing interests.
References
- Mark Welch JL, Rossetti BJ, Rieken CW, et al. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U. S. A. 113, E791–E800 (2016).
- Dupuy AK, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9, e90899–e90899 (2014).
- Chen T, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010, baq013–baq013 (2010).
- Fakhruddin KS, Ngo HC & Samaranayake LP. Cariogenic microbiome and microbiota of the early primary dentition: A contemporary overview. Oral Dis. 25, 982–995 (2019).
- Tanner ACR, Kressirer CA, Rothmiller S, et al. The Caries Microbiome: Implications for Reversing Dysbiosis. Adv. Dent. Res. 29, 78–85 (2018).
- Kilian M, et al. The oral microbiome – an update for oral healthcare professionals. Br. Dent. J. 221, 657–666 (2016).
- Hall MW, et al. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ biofilms microbiomes 3, 2 (2017).
- Zhou Y, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 14, R1–R1 (2013).
- Wright CJ, et al. Microbial interactions in building of communities. Mol. Oral Microbiol. 28, 83–101 (2013).
- Baraniya D, et al. Supragingival mycobiome and inter-kingdom interactions in dental caries. J. Oral Microbiol. 12, 1729305 (2020).
- Garmaeva S, et al. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol. 17, 84 (2019).
- Sztajer H, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271 (2014).
- Eidelman E, Faibis S & Peretz BA comparison of restorations for children with early childhood caries treated under general anesthesia or conscious sedation. Pediatr. Dent. 22, 33–7 (2000).
- Bramhachari PV, Ahmed VKS, Selvin J, et al. Quorum Sensing and Biofilm Formation by Oral Pathogenic Microbes in the Dental Plaques: Implication for Health and Disease BT - Implication of Quorum Sensing and Biofilm Formation in Medicine, Agriculture and Food Industry. in (ed. Bramhachari, P. V.) 129–140 (Springer Singapore, 2019). doi:10.1007/978-981-32-9409-7_10.
- Bryan NS, Tribble G & Angelov N. Oral Microbiome and Nitric Oxide: the Missing Link in the Management of Blood Pressure. Curr. Hypertens. Rep. 19, 33 (2017).
- Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, R42–R42 (2012).
- Krom BP, Kidwai S & ten Cate JM. Candida and Other Fungal Species: Forgotten Players of Healthy Oral Microbiota. J. Dent. Res. 93, 445–451 (2014).
- Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6, e1000713–e1000713 (2010).
- Ursell LK, Metcalf JL, Parfrey LW, et al. Defining the human microbiome. Nutr. Rev. 70 Suppl 1, S38–S44 (2012).
- Kianoush N, et al. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS One 9, e92940–e92940 (2014).
- Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013).
- Goulas T, et al. A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome. J. Biol. Chem. 292, 10883–10898 (2017).
- Koopman JE, et al. Stability and Resilience of Oral Microcosms Toward Acidification and Candida Outgrowth by Arginine Supplementation. Microb. Ecol. 69, 422–433 (2015).
- Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6, 915–926 (2012).
- Wade WG. Characterisation of the human oral microbiome. J. Oral Biosci. 55, 143–148 (2013).
- Dame-Teixeira N, et al. Presence of Archaea in dental caries biofilms. Arch. Oral Biol. 110, 104606 (2020).
- Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732 (2005).
- Collins MD & Lawson PA. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int. J. Syst. Evol. Microbiol. 50, 365–369 (2000).
- Featherstone JDB. The Science and Practice of Caries Prevention. J. Am. Dent. Assoc. 131, 887–899 (2000).
- Reynolds EC. Calcium phosphate-based remineralization systems: scientific evidence? Aust. Dent. J. 53, 268–273 (2008).
- Featherstone JD. Caries prevention and reversal based on the caries balance. Pediatr Dent. 2006 Mar-Apr;28(2):128-32; discussion 192-8. PMID: 16708787.
- Ledder RG, Kampoo K, Teanpaisan R, et al. Oral Microbiota in Severe Early Childhood Caries in Thai Children and Their Families: A Pilot Study. Front. Microbiol. 9, 1–10 (2018).
- Aas JA, et al. Bacteria of Dental Caries in Primary and Permanent Teeth in Children and Young Adults. J. Clin. Microbiol. 46, 1407 LP – 1417 (2008).
- Gross EL, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48, 4121–4128 (2010).
- Li Y, Ge Y, Saxena D, et al. Genetic Profiling of the Oral Microbiota Associated with Severe Early-Childhood Caries. J. Clin. Microbiol. 45, 81 LP – 87 (2007).
- Dalmasso M, et al. Isolation of a Novel Phage with Activity against Streptococcus mutans Biofilms. PLoS One 10, e0138651–e0138651 (2015).
- Li Y, Caufield PW, Dasanayake AP, et al. Mode of Delivery and Other Maternal Factors Influence the Acquisition of Streptococcus mutans in Infants. J. Dent. Res. 84, 806–811 (2005).
- Poklepovic Pericic T, et al. WITHDRAWN: Interdental brushing for the prevention and control of periodontal diseases and dental caries in adults. Cochrane database Syst. Rev. 4, CD009857–CD009857 (2019).
- Albino J & Tiwari T. Preventing Childhood Caries: A Review of Recent Behavioral Research. J. Dent. Res. 95, 35–42 (2016).
- Griffin SO, Regnier E, Griffin PM, et al. Effectiveness of Fluoride in Preventing Caries in Adults. J. Dent. Res. 86, 410–415 (2007).
- Gooch BF, Griffin SO & Malvitz DM. The Role of Evidence in Formulating Public Health Programs to Prevent Oral Disease and Promote Oral Health in the United States. J. Evid. Based Dent. Pract. 6, 85–89 (2006).
- Warinner C, et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 46, 336–344 (2014).
- Adler CJ, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 45, 450–455 (2013).
- Ziesemer KA, et al. Intrinsic challenges in ancient microbiome reconstruction using 16S rRNA gene amplification. Scientific reports vol. 5 16498 (2015).
- Olsen I & Yamazaki K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 11, 1586422 (2019).
- Jones BV & Gahan CGM. Metagenomic Analysis of Bile Salt Hydrolases in the Human Gut Microbiome BT - Encyclopedia of Metagenomics. in (ed. Nelson, K. E.) 1–13 (Springer New York, 2013). doi:10.1007/978-1-4614-6418-1_777-1.
- McDonald JE, et al. Characterising the Canine Oral Microbiome by Direct Sequencing of Reverse-Transcribed rRNA Molecules. PLoS One 11, e0157046 (2016).
- Hodkinson BP & Grice EA. Next-Generation Sequencing: A Review of Technologies and Tools for Wound Microbiome Research. Adv. wound care 4, 50–58 (2015).
- Goodwin S, McPherson JD & McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
- Ghanbari M, Kneifel W & Domig, KJA. new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 448, 464–475 (2015).
- Walsh CJ, Guinane CM, O’ Toole PW, et al. A Profile Hidden Markov Model to investigate the distribution and frequency of LanB-encoding lantibiotic modification genes in the human oral and gut microbiome. PeerJ 5, e3254–e3254 (2017).
- Kolenbrander PE, et al. Bacterial interactions and successions during plaque development. Periodontol. 2000 42, 47–79 (2006).
- Kolenbrander PE. Oral Microbial Communities: Biofilms, Interactions, and Genetic Systems. Annu. Rev. Microbiol. 54, 413–437 (2000).
- D’Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 17, 254–264 (2010).
- Nobbs AH, Jenkinson HF & Jakubovics NS. Stick to your gums: mechanisms of oral microbial adherence. J. Dent. Res. 90, 1271–1278 (2011).
- Lamont RJ, Koo H & Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759 (2018).
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
- Xiao J, Fiscella KA & Gill SR. Oral microbiome: possible harbinger for children’s health. Int. J. Oral Sci. 12, 12 (2020).
- Graves DT, Corrêa JD & Silva TA. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 98, 148–156 (2018).
- Fong IW. Emerging relations between infectious diseases and coronary artery disease and atherosclerosis. CMAJ 163, 49–56 (2000).
- Demmer RT & Desvarieux M. Periodontal infections and cardiovascular disease: The heart of the matter. J. Am. Dent. Assoc. 137, S14–S20 (2006).
- Schmidt TSB, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife 8, 8–10 (2019).
- Gao L, et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell 9, 488–500 (2018).
- Bartlett A, Gullickson RG, Singh R, et al. The link between oral and gut microbiota in inflammatory bowel disease and a synopsis of potential salivary biomarkers. Appl. Sci. 10, 1–22 (2020).
- Pflughoeft KJ & Versalovic J. Human Microbiome in Health and Disease. Annu. Rev. Pathol. Mech. Dis. 7, 99–122 (2012).
- Filoche S, Wong L & Sissons CH. Oral Biofilms: Emerging Concepts in Microbial Ecology. J. Dent. Res. 89, 8–18 (2009).
- Kalpana B, et al. Bacterial diversity and functional analysis of severe early childhood caries and recurrence in India. Sci. Rep. 10, 21248 (2020).
- Avila M, Ojcius DM & Yilmaz Ö. The Oral Microbiota: Living with a Permanent Guest. DNA Cell Biol. 28, 405–411 (2009).
- Turnbaugh PJ, et al. The Human Microbiome Project. Nature 449, 804–810 (2007).
- Abusleme L, et al. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI insight vol. 3 (2018).
- Hwang G, Marsh G, Gao L, et al. Binding Force Dynamics of Streptococcus mutans-glucosyltransferase B to Candida albicans. J. Dent. Res. 94, 1310–1317 (2015).
- Liu W, et al. Deciphering links between bacterial interactions and spatial organization in multispecies biofilms. ISME J. 13, 3054–3066 (2019).
- Cavalcanti IMG, Nobbs AH, Ricomini-Filho AP, et al. Interkingdom cooperation between Candida albicans, Streptococcus oralis and Actinomyces oris modulates early biofilm development on denture material. Pathog. Dis. 74, (2016).
- Diaz PI, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80, 620–632 (2012).
- Cephas KD, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One 6, e23503–e23503 (2011).
- Bor B, Cen L, Agnello M, et al. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci. Rep. 6, 27956 (2016).
- Falsetta ML, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82, 1968–1981 (2014).
- Koo H, Xiao J, Klein MI, et al. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192, 3024–3032 (2010).
- Soro V, et al. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl. Environ. Microbiol. 80, 6480–6489 (2014).
- Kuehbacher T, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med. Microbiol. 57, 1569–1576 (2008).
- Xu X, et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ. Microbiol. 17, 699–710 (2015).
- Costalonga M & Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 162, 22–38 (2014).
- Abusleme L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025 (2013).
- Ly M, et al. Altered Oral Viral Ecology in Association with Periodontal Disease. MBio 5, e01133-14 (2014).
- Mukherjee PK, et al. Oral Mycobiome Analysis of HIV-Infected Patients: Identification of Pichia as an Antagonist of Opportunistic Fungi. PLOS Pathog. 10, e1003996 (2014).
- Minot S, et al. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. 110, 12450 LP – 12455 (2013).
- Willner D, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc. Natl. Acad. Sci. U. S. A. 108 Suppl, 4547–4553 (2011).
- Baker JL, Bor B, Agnello M, et al. Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol. 25, 362–374 (2017).
- Qi Y, et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics (2020) doi: https://doi.org/10.1016/j.ygeno.2020.09.063.
