Leflunomide Confers Rapid Recovery from COVID-19 and is Coupled with Temporal Immunologic Changes
Ada Alice Dona1,2#, James F Sanchez1#, Joycelynne M Palmer3, Timothy W. Synold4, Flavia Chiuppesi5, Sandra Thomas2, Enrico Caserta1,2, Mahmoud Singer1,2, Theophilus Tandoh1,2, Arnab Chowdhury3, Amrita Krishnan1, Michael Rosenzweig1, Don J Diamond5, Steven Rosen1*, Flavia Pichiorri1,2*, Sanjeet Dadwal6*
1Judy and Bernard Briskin Center for Multiple Myeloma Research, Department of Hematology and Hematopoietic Cell Transplantation, City of Hope, Duarte, CA
2Department of Hematologic Malignancies Translational Science, Beckman Research Institute, City of Hope, Duarte, CA
3Department of Computational and Quantitative Sciences, Beckman Research Institute, City of Hope, Duarte, CA
4Department of Cancer Biology, City of Hope, Duarte, CA
5Department of Experimental Therapeutics, City of Hope, Duarte, CA
6Department of Medicine, Division of Infectious Disease, City of Hope, Duarte, CA USA
#These authors contributed equally to this work.
Abstract
Background: Vaccines for SARS-CoV-2 have been considerably effective in reducing rates of infection and severe COVID-19. However, many patients, especially those who are immunocompromised due to cancer or other factors, as well as individuals who are unable to receive vaccines or are in resource-poor countries, will continue to be at risk for COVID-19. We describe clinical, therapeutic, and immunologic correlatives in two patients with cancer and severe COVID-19 who were treated with leflunomide after failing to respond to standard-of-care comprising remdesivir and dexamethasone. Both patients had breast cancer and were on therapy for the malignancy.
Methods: The protocol is designed with the primary objective to assess the safety and tolerability of leflunomide in treating severe COVID-19 in patients with cancer. Leflunomide dosing consisted of a loading dose of 100 mg daily for the first three days, followed by daily dosing, at the assigned dose level (Dose Level 1: 40 mg, Dose Level -1, 20 mg; Dose Level 2, 60 mg), for an additional 11 days. At defined intervals, serial monitoring of blood samples for toxicity, pharmacokinetics, and immunologic correlative studies were performed, as well as nasopharyngeal swabs for PCR analysis of SARS-CoV-2.
Results: Preclinically, leflunomide impaired viral RNA replication, and clinically, it led to a rapid improvement in the two patients discussed herein. Both patients completely recovered, with minimal toxicities; all adverse events experienced were considered unrelated to leflunomide. Single-cell mass-cytometry analysis showed that leflunomide increased levels of CD8+ cytotoxic and terminal effector T cells and decreased naïve and memory B cells.
Conclusions: With ongoing COVID-19 transmission and occurrence of breakthrough infections in vaccinated individuals, including patients with cancer, therapeutic agents that target both the virus and host inflammatory response would be helpful despite the availability of currently approved anti-viral agents. Furthermore, from an access to care perspective, especially in resource-limited areas, an inexpensive, readily available, effective drug with existing safety data in humans is relevant in the real-world setting.
Background
Vaccines for SARS-CoV-2, the causative agent of COVID-19, have been considerably effective in reducing rates of infection and severe COVID-19. However, many individuals, including some patients with cancer, people who are unable to receive vaccines, or individuals in resource-poor countries, where access to antiviral medications, monoclonal antibodies, and vaccines is limited, will continue to be at risk for COVID-19. In infected patients, the non-specific innate immune response is followed by adaptive immunity mediated by both B cells and T cells,1 but patients with cancer and other disorders may be impacted by a dysregulated immune response.
Leflunomide is an FDA-approved, oral agent that has been commercially available since 1998. It is used for the treatment of rheumatoid arthritis as a single agent or in combination with methotrexate. Its primary mechanism of action is inhibiting de novo pyrimidine synthesis by targeting dihydroorotate dehydrogenase (DHODH)2, with an anti-proliferative effect in B- and T-lymphocytes3. Teriflunomide is the active primary metabolite of leflunomide.
Host-targeting antiviral drugs may be appealing by targeting the host machinery exploited by the virus, thereby potentially applying to a wide range of viruses and viral strains. To this end, repurposed drugs are especially suitable, as many of the extensive preclinical validation and toxicity studies have already been performed. As a DHODH inhibitor, leflunomide is worthy of study for SARS-CoV-2, an RNA virus, particularly because of the virus’s high content of uracil4, one of the two nucleobases inhibited by leflunomide. Leflunomide has been used in difficult-to-treat cytomegalovirus infections in patients undergoing hematopoietic cell transplantation and in polyoma virus (BK virus) hemorrhagic cystitis when no alternatives are available.
Methods
The protocol is designed as a phase 1/ 2 trial with the primary objective to assess the safety and tolerability of leflunomide in treating severe COVID-19 in patients with cancer (NCT04532372). The study was performed in accordance with the provisions of the Declaration of Helsinki and approved by the City of Hope Institutional Review Board. All participants gave written informed consent. Leflunomide dosing comprised a loading dose of 100 mg daily for the first three days, followed by daily dosing at the assigned dose level (Dose Level 1: 40 mg, Dose Level -1, 20 mg; Dose Level 2, 60 mg), for an additional 11 days. At defined intervals, serial monitoring of blood samples for toxicity, pharmacokinetics, and immunologic correlative studies were performed, as well as nasopharyngeal swabs for PCR analysis of SARS-CoV-2.
Primary Samples
Biospecimen collection and immunological analyses are detailed below. Briefly, peripheral blood from healthy donors was obtained from the institutional hematopoietic tissue repository. Specifically, the cellular fraction of the peripheral blood mononuclear cells (PBMCs) was isolated using Ficoll-Paque Plus (GE Healthcare) following the manufacturer’s instructions.
Mass Cytometry (CyTOF) Staining and Acquisition
A total of 2-4x106 PBMCs were stained with a panel containing 30 metal-conjugated antibodies (Sup. Table 1) according to Fluidigm's protocol for Maxpar Antibody Labeling (PRD002 Rev 12). PBMCs derived from the clinical trial were stained with Fluidigm's Maxpar Direct Immune Profiling Assay Cell Staining (PN 400286 B1). Samples were acquired, exported as FCS files, and normalized on Fluidigm's Helios (Software 7.0.5189).
Supplementary Table 1: Maxpar direct immune profiling assay 30-marker panel with clones and heavy metals (Fluidigm)
|
Target |
Clone |
Metal |
|
Anti-human CD45 |
HI30 |
89Y |
|
Live/dead 103Rh-Intercalator (500 μM) |
N/A |
103Rh |
|
Anti-human CD196/CCR6 |
G034E3 |
141Pr |
|
Anti-human CD123 |
6H6 |
143Nd |
|
Anti-human CD19 |
HIB19 |
144Nd |
|
Anti-human CD4 |
RPA-T4 |
145Nd |
|
Anti-human CD8a |
RPA-T8 |
146Nd |
|
Anti-human CD11c |
Bu15 |
147Sm |
|
Anti-human CD16 |
3G8 |
148Nd |
|
Anti-human CD45RO |
UCHL1 |
149Sm |
|
Anti-human CD45RA |
HI100 |
150Nd |
|
Anti-human CD161 |
HP-3G10 |
151Eu |
|
Anti-human CD194/CCR4 |
L291H4 |
152Sm |
|
Anti-human CD25 |
BC96 |
153Eu |
|
Anti-human CD27 |
O323 |
154Sm |
|
Anti-human CD57 |
HCD57 |
155Gd |
|
Anti-human CD183/CXCR3 |
G025H7 |
156Gd |
|
Anti-human CD185/CXCR5 |
J252D4 |
158Gd |
|
Anti-human CD28 |
CD28.2 |
160Gd |
|
Anti-human CD38 |
HB-7 |
161Dy |
|
Anti-human CD56/NCAM |
NCAM16.2 |
163Dy |
|
Anti-human TCRgd |
B1 |
164Dy |
|
Anti-human CD294 |
BM16 |
166Er |
|
Anti-human CD197/CCR7 |
G043H7 |
167Er |
|
Anti-human CD14 |
63D3 |
168Er |
|
Anti-human CD3 |
UCHT1 |
170Er |
|
Anti-human CD20 |
2H7 |
171Yb |
|
Anti-human CD66b |
G10F5 |
172Yb |
|
Anti-human HLA-DR |
LN3 |
173Yb |
|
Anti-human IgD |
IA6-2 |
174Yb |
|
Anti-human CD127 |
A019D5 |
176Yb |
CyTOF Analysis
Non-custom panel analysis was analyzed using Maxpar Pathsetter™ software powered by GemStone 2.0.41, Verity Software House, Topsham, Maine (Version 2.0.45). The FCS files were also analyzed using FlowJo™ Software (Windows edition, Version 10.6. Becton Dickinson Company; 2019), and the Cytobank© platform (https://www.cytobank.org) (Cytobank, Inc., Mountain View, CA) was used for gating, tSNE plotting, and FlowSOM.
Pharmacokinetic Analysis
Pharmacokinetic sampling was performed per protocol during active treatment with leflunomide. Samples were collected prior to initiation of drug administration, daily for days +1 to +7 of treatment, and subsequently daily if the subject was hospitalized, or during visits if the subject was outpatient. Plasma was analyzed for total (bound and unbound) and free (unbound) teriflunomide concentrations using a previously described LC-MS/MS method5. Briefly, following addition of a deuterated teriflunomide internal standard, plasma (total drug) or ultrafiltered plasma (free drug) samples were prepared by protein precipitation using 3:1 ice cold methanol. The resulting protein-free plasma or ultrafiltered plasma was further diluted 10,000-fold with 0.5 mM ammonium acetate, 0.025% formic acid in 75% methanol, and 10 µl was injected for analysis. The lower limits of quantitation of the assay were 30 µg/ml and 30 ng/ml in plasma and ultrafiltered plasma, respectively.
DLT Definitions
Dose limiting toxicity (DLT) was defined as any of the following toxicities that were at least possibly related to leflunomide:
- Hematologic DLT events (any adverse events [AEs] considered at least possibly attributable to study treatment):
- For patients with solid tumor malignancies, all grade 3 or 4 hematologic AEs
- For patients with hematologic malignancies, the following AEs would be considered DLTs if they were determined as not attributable to underlying hematological malignancy.
- Grade 4 neutropenia (absolute neutrophil count [ANC] <500/mm3)
- Grade 3 or 4 febrile neutropenia
- Grade 4 thrombocytopenia (<25,000/mm3)
- Grade 3 thrombocytopenia (<50,000/mm3) with bleeding
- Non-Hematologic AEs (any treatment emergent AEs):
- ≥ Grade 3 non-hematologic toxicity excepting the following:
- Alopecia
- Grade 3 nausea/vomiting/diarrhea for less than 72 hours treated with adequate antiemetic and other supportive care
- Grade 3 fatigue for < 1 week
- ≥ Grade 3 electrolyte abnormalities that are not clinically complicated and resolve spontaneously or to conventional medical interventions within 72 hours
- ≥ Grade 3 amylase or lipase elevation not associated with symptoms or clinical manifestations of pancreatitis
- Treatment-emergent increase in serum ALT or AST to > 3x ULN associated with an increase in serum total bilirubin to > 2x ULN (consistent with Hy’s Law).
- Any treatment-emergent Grade 5 AE that occurs during the 28-day treatment period that is not due to underlying malignancy would be considered a DLT.
Cholestyramine
Leflunomide is reabsorbed from the gastrointestinal tract and consequently is detectable in the body for up to 2 years. Cholestyramine, a bile acid sequestrant, fixes the metabolite, preventing reabsorption and expediting drug elimination. We administered cholestyramine daily starting on day 29 until plasma levels of teriflunomide were less than 0.02 mg/L by two separate measurements 14 days apart.
Cell culture
Multiple myeloma (MM) cell lines MM.1S and U266 were purchased from ATCC. All cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) (Cat. #019K8420, Sigma), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Cat.#15140-122, Gibco).
Immunoblotting
Cells lyses in RIPA buffer (89901, Thermo Scientific) were supplemented with protease and phosphatase inhibitors, and then sonicated (30” Pulse ON 02” Pulse OFF 03” 60% amplification). Lysates were then clarified by spinning at 14,000 rpm at 4°C and protein concentration quantified by BCA Protein Assay (23227, Thermo Scientific). Forty micrograms of proteins were denatured in boiling SDS sample buffer, resolved on 4%-20% gradient gels (Cat.# 5671093, Bio-Rad), and transferred to nitrocellulose membranes (Cat.# 1704271 Bio-Rad). After blocking nonspecific binding of antibody with 5% BSA (Fisher BioReagents), blots were probed with anti-RV σ-NS (1:1000; provided by Oncolytics Biotech Inc.) to assess viral replication for σ1 protein, or GAPDH (Cat.# sc-32233, Santa Cruz Biotechnology) as internal control. Blots were washed three times for 15 minutes with TBST 1X and stained with horseradish peroxidase (HRP)-conjugated secondary antibodies (diluted 1:4000) for 2hrs at room temperature. Primary antibodies were detected by binding with secondary antibodies donkey anti-goat IgG (H+L) (Cat.# A16005, Invitrogen) and goat anti-mouse IgG-HRP (NA931, GE Healthcare), and using an enhanced chemiluminescent visualization system (Cat.#RPN2209 ECL Western Blotting Detection Reagents, GE Healthcare). Primary and secondary antibodies were diluted according to the manufacturer instructions. The bands were quantified by densitometry analyses using Image Lab program (Biorad) and normalized to GAPDH.
RNA isolation and analysis
Total cellular RNA was extracted by using TRIZOL reagent (Cat. #15596018 Invitrogen Corporation) and RNA Clean-Up and Concentration Kit (Cat. #43200 Norgen) according to the manufacturer’s protocols. cDNA synthesis was performed by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Cat# 4368814). Reverse transcription reactions were run using a Mastercycler pro. Quantitative real time-PCR (qRT-PCR) was performed with the TaqMan method (Applied Biosystems), according to the manufacturer’s instructions. The appropriate TaqMan probes for mRNA quantification were purchased from Applied Biosystems, and all reactions were performed in triplicate. The following probes were used: (HS99999905_m1) GAPDH used as endogenous control; (HS00989291_m1) INF-γ; (Hs00961622 m1) IL-10; (Hs00174131 m1) IL-6; (Hs01077958 S1) IFNB1; (Hs00174128 m1) TNFalpha; (Hs00265051 S1) IFNA2.
For the quantification of viral RNA of genomes extracted from infected PBMCs from healthy donors and MM cell lines, q-RT-PCR reactions were conducted using the PowerUp SYBR Green Master Mix (Applied Biosystems Cat.# 4367659) according to the manufacturer’s instructions and the following primers: Reo9: 5′-TG CGC AAG AGG CAG CAA TCG-3′ and Reo10: 5′-TT CGC GGG CCT CGC ACA TTC-3′; GAPDH FD: 5′- CTG CAC CAC CAA CTG CTT -3′ and GAPDH RV: 5′- CAT GAC GGC AGG TCA GGT -3′.
Cytokine measurements
Samples were analyzed for 30 cytokines using the Human Cytokine Thirty-Plex Antibody Magnetic Bead Kit (Invitrogen, Camarillo, CA) as per the manufacturer’s protocol. Brieï¬y, Invitrogen’s multiplex bead solution was vortexed for 30 seconds, and 25 µl was added to each well of a flat-bottom 96 well microplate. Samples (e.g., plasma) were diluted 1:2 with assay diluent and loaded into the wells containing 50 µl of incubation buffer. Cytokine standards were reconstituted with assay diluent, and serial dilutions of cytokine standards were prepared in parallel and added to the plate. Plates were incubated on a shaker at 500 rpm in the dark at room temperature for 2 hours. The plate was then applied to a magnetic capture device and washed three times with 200 µl of wash buffer. After the final wash, 200 µl of a biotinylated detection antibody mixture was added to each well, and the plate was incubated on a shaker for 1 hour. After washing again three times with 200 µl of wash buffer, streptavidin-phycoerythrin (100 µl) was added to the wells. The plate was incubated on a plate shaker for another 30 minutes and washed three times, after which the beads were resuspended in 150 µl of wash buffer and shaken for 1 minute. Finally, the assay plate was transferred to a Flexmap 3D Luminex system (Luminex Corp, Austin, TX) for analysis. Cytokine concentrations were calculated using Bio-Plex Manager 6.0 software with a ï¬ve parameter curve-ï¬tting algorithm applied for standard curve calculations for duplicate samples.
Results and Discussion
Previous data have demonstrated the preclinical anti-viral activity of DHODH inhibitors against RNA viruses such as influenza, Ebola, and Zika viruses, as well as SARS-CoV-26. In support of the importance of DHODH in viral replication, viral growth was largely inhibited in DHODH knockout cells, and addition of uracil and cytosine (the pyrimidine bases) restored viral activity6.
Reovirus serotype 3–dearing strain (RV) is a naturally occurring, ubiquitous, nonenveloped human reovirus with a genome that consists of 10 segments of double-stranded RNA. As with SARS-CoV-2, RV can infect human and animals and never passes through a DNA phase. It actively replicates its RNA genome and induces a strong anti-viral immune response. The two viruses primarily differ in the composition of the envelope and the fact that coronaviruses need to synthetize the RNA negative strand before initiating transcription. Our preliminary data show that leflunomide significantly arrested RV productive infection in cancer cells, as shown by decreased capsid formation and genome replication (Sup. Fig. 1A-F). Impairment in viral replication was also observed when PBMCs were treated with teriflunomide, the active metabolite of leflunomide, in combination with RV, compared to the virus alone (Sup. Fig. 1G-H). Our data also show that, in the ex vivo setting, the addition of teriflunomide to RV-infected PBMCs significantly decreased the expression of highly inflammatory cytokines such as IL-6 and GM-CSF (Sup. Fig. 1I-J) but concomitantly enhanced the anti-viral interferon I (IFN-α and IFN-β) response and TNF-α (Sup. Fig. 1K-L-M). The antiviral activity of leflunomide aligns with the experience of the drug as an agent against cytomegalovirus (CMV) and polyoma BK virus infections in immunocompromised hosts7,8.

Supplementary Figure 1: A-B-C)Western Blot analysis of σ-NS viral protein in U266 cells after 6, 16 and 48hrs of treatment with teriflunomide (100 µm) and/or RV (5 MOI) infection (A), and q-RT-PCR for the viral genome expression after 16hrs (B) or 48hrs (C), normalized compared to control GAPDH and expressed as the mean ± SEM of triplicates in fold-change compared to the control; D-E-F) Western Blot analysis of σ-NS viral protein in MM1.S cells after 16 and 48hrs of treatment with teriflunomide (100 µm) and/or RV (5 MOI) infection (D), and q-RT-PCR for the viral genome expression after 16hrs (E) or 48hrs (F), normalized compared to control GAPDH and expressed as the mean ± SEM of triplicates in fold-change compared to the control; G-H) Western Blot analysis of (σ-NS) viral protein in healthy donor PBMC treated for 48hrs with teriflunomide (100 µM) and infected with RV (5 MOI); H) After 16hrs, RNA was isolated, and expression of the RV genome was determined by q-RT-PCR. Data were normalized compared to control GAPDH and expressed as the mean ± SEM of triplicates as fold-change compared to the control; I-J-K-L-M) PBMCs obtained from healthy donors were treated with teriflunomide (100 µM) and/or RV (5 MOI) and infected for 16hrs to assess the cytokine (IL-6, GM-CSF, IFN-α, IFN-β, TNF-α) mRNA expression profile by q-RT-PCR. For IL-6 and GM-CSF, data are expressed as the mean ± SEM (IL-6 and GM-CSF n=2 healthy donors; IFN-α, IFN-β and TNF-α n=3 healthy doors), normalized compared to control GAPDH; Comparisons among groups were performed by one-way ANOVA.
We furthermore describe our findings of two patients with breast cancer who were admitted to our COVID unit after worsening shortness of breath due to COVID-19. Both patients’ oxygen levels decreased to below 90%, requiring high-flow oxygen. Computed tomography (CT) of the chest revealed multifocal pneumonia in both patients (Fig. 1A-B). The patients began treatment with remdesivir, dexamethasone, cefepime, and azithromycin, with minimal improvement. No co-infections were identified, and the clinical condition was due to COVID-19.
We administered leflunomide to both patients following informed consent and confirmation of eligibility to participate in the clinical trial. After loading with leflunomide with a dose of 100 mg daily x 3 days, the patients received 40 mg daily for 11 additional days. Leflunomide is reabsorbed from the gastrointestinal tract and consequently is detectable in the body for up to 2 years. Cholestyramine, a bile acid sequestrant, fixes the metabolite, preventing reabsorption and expediting drug elimination. We administered cholestyramine daily starting on day 29 until plasma levels of teriflunomide were less than 0.02 mg/L by two separate measurements 14 days apart. Plasma concentrations of teriflunomide were also measured throughout the treatment period in both patients as previously described9. Maximum plasma concentrations (Cmax) achieved in the two subjects were 252.4 and 152.3 µmol/L, respectively, and the areas-under-the-curve (AUC) were 5598.0 and 3498.1 µmol/L x day. As shown in the concentration-versus-time plots (Sup. Fig. 2A), plasma concentrations were maintained above the in vitro EC50 against SARS-CoV-2 (26 µmol/L) 6 for >36 days.
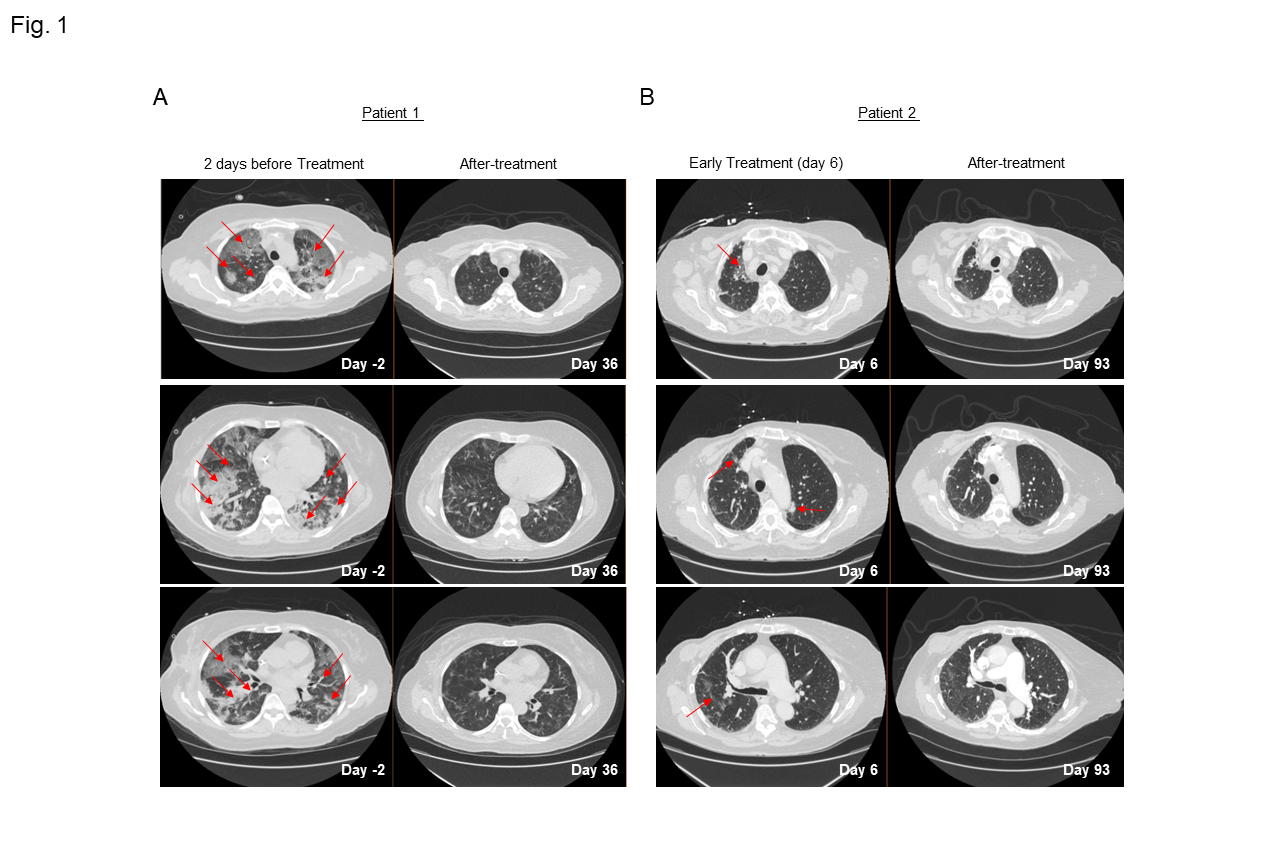
Figure 1: A-B)Serial CT chest scans from patient 1 (A) and 2 (B) with severe COVID-19 pneumonia showing peripheral multilobar ground glass opacities (red arrow) two days before the initiation of leflunomide treatment for patient 1 and Day 6 of treatment for patient 2. Scans obtained after 36 days (A) and after 93 days (B) on treatment show partial absorption of the abnormalities for both patients.
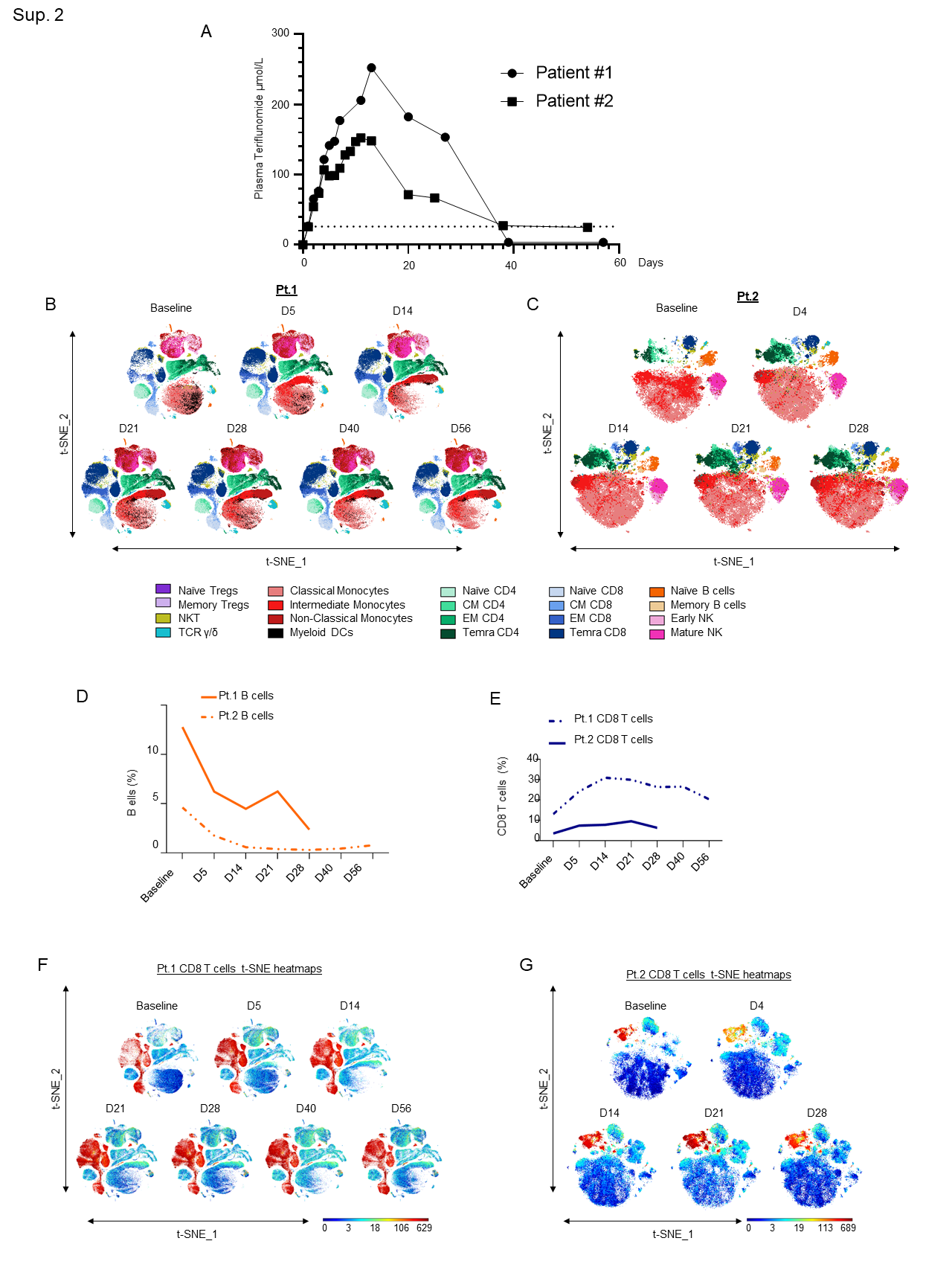
Supplementary Figure 2: A)Teriflunomide plasma concentration-versus-time plots for 2 patients receiving leflunomide. The dashed line represents the in vitro EC50 for teriflunomide against SARS-CoV-2 (26 µmol/L). B-C) In-depth longitudinal immune profiling of two patients with COVIDâ19 and breast cancer (A and B) treated with 40 mg leflunomide. A Maxpar Direct Immune Profiling System using a dry 30-marker antibody panel was employed. Hierarchical clustering and statistical mapping were performed algorithmically via the Cytobank© platform. vi-SNE analysis (iterations=1000, perplexity=100) are displayed in 2D plots using the resultant t-SNE 1 and t-SNE2 dimensions. High-fidelity FlowSOM (“self-organizing map”) (metacluster=10 and cluster=100) based on vi-SNE 2D plots showing 20 different immune-compartments was used; D) Line graphs showing B cell expression as a percentage of the total PBMC population in Patients 1 and 2 as measured by CyTOF; E) Line graphs showing CD8+ T cell expression as a percentage of the total PBMC population in Patients 1 and 2 as measured by CyTOF; F-G) t-SNE heatmaps showing overall CD8+ T cell expression in both patients.
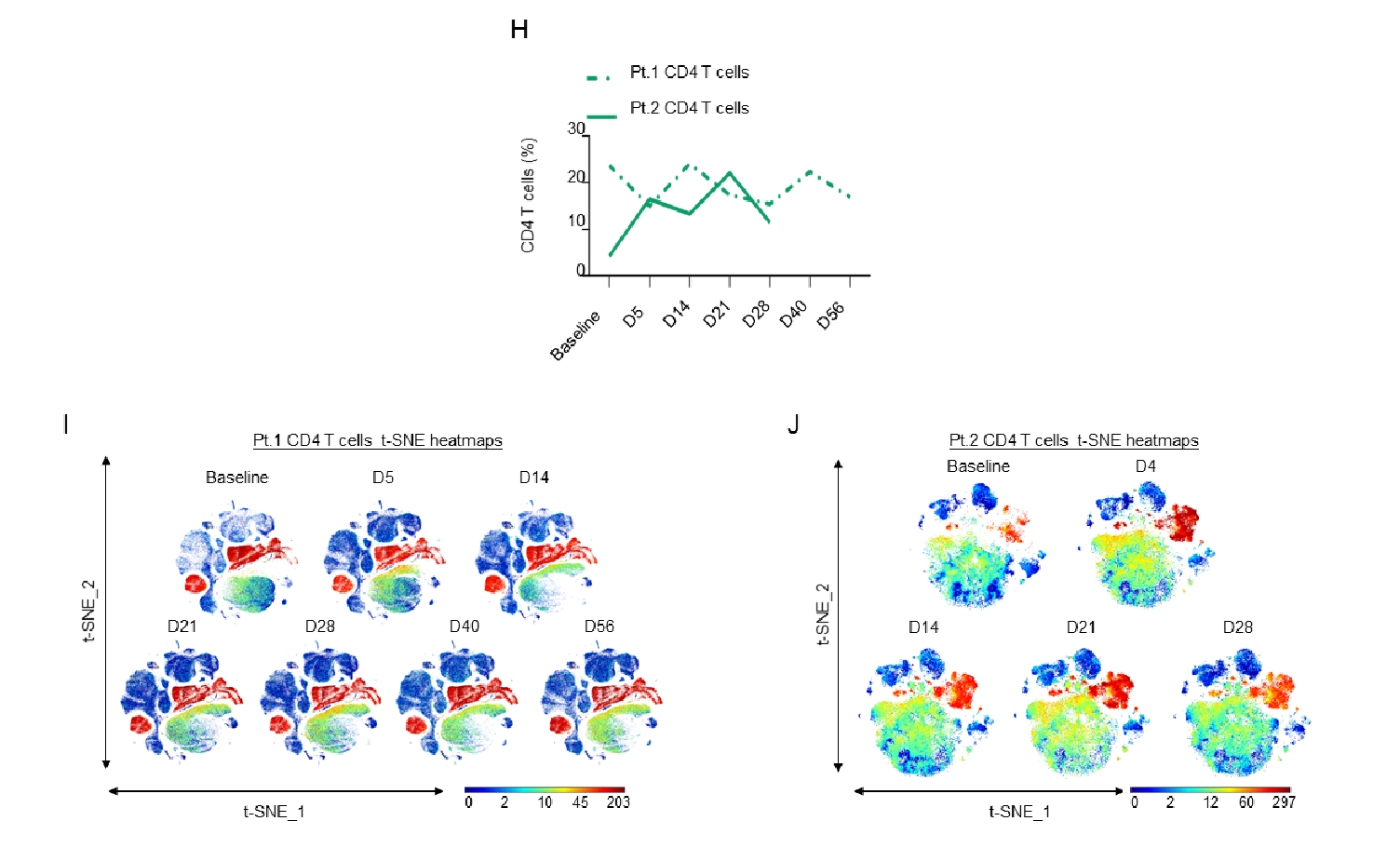
Supplementary Figure 2: H)Line graphs showing CD4+ T cell expression as a percentage of the total PBMC population in Patients 1 and 2 as measured by CyTOF; I-J) t-SNE heatmaps showing overall CD4+ T cell expression in both patients.
The patients had rapid improvement in symptoms and oxygenation and experienced a complete recovery as clearly shown from the CT scans of the lung before and after combination therapy that included leflunomide (Fig. 1A-B). The adverse events that were observed were minimal and considered unrelated to leflunomide. These findings are consistent with a pilot study in which 15 patients given leflunomide had improved viral clearance and hospital discharge rate when compared to 12 patients in a control arm10. Because leflunomide is an immune suppressive drug, which could potentially impair its anti-viral activity, we decided to follow the immune changes of these patients during and after the course of treatment. A 30-marker single cell mass cytometry (CyTOF) antibody panel (Sup. Table 1) designed to identify twenty different immune compartments (Sup. Table 2) was used to perform a longitudinal high dimensional immune profiling (Fig. 2A and Sup. Fig. 2B-C).
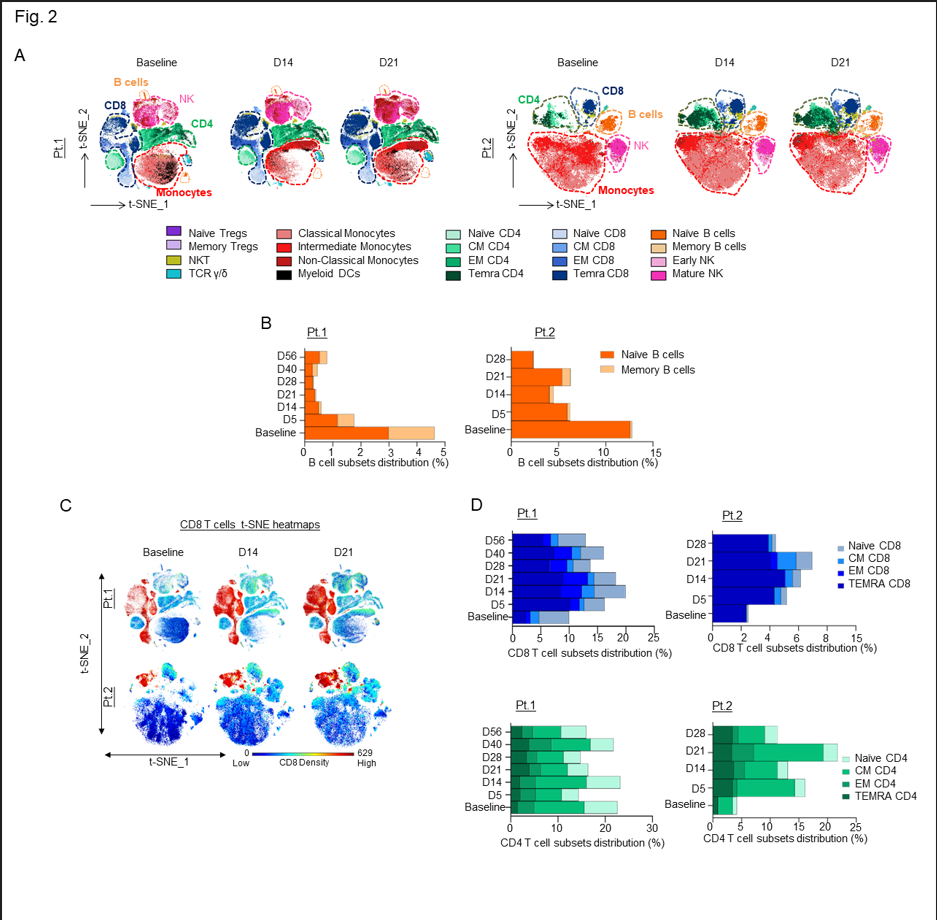
Figure 2: A) In-depth longitudinal immune profiling of two patients with COVIDâ19 and breast cancer treated with 40 mg leflunomide. A Maxpar Direct Immune Profiling System using a dry 30-marker antibody panel was employed. Hierarchical clustering and statistical mapping were performed algorithmically via the Cytobank© platform. vi-SNE analysis (iterations=1000, perplexity=100) are displayed in 2D plots using the resultant t-SNE 1 and t-SNE 2 dimensions. High-fidelity FlowSOM (“self-organizing map”) (metacluster=10 and cluster=100) based on vi-SNE 2D plots showing 20 different immune-compartments was used; B) Bar graphs representing the expression of naïve and memory B cell subpopulations through the course of treatment; C) t-SNE heatmap highlighting the increased expression of CD8+ cytotoxic T cells at baseline and on days 14 and 21 of treatment in both patients; D) Bar graphs showing effector memory and terminally differentiated effector T cells (TEMRA) T cell subsets in percentages compared to the total count PBMCs in both patients.
Supplementary Table 2: CyTOF gating strategy
|
Cell subsets |
Model phenotypes |
|
Naïve B-cells |
CD45+ CD3- CD19+ CD20+ HLA-DR+ CD27- |
|
Memory B-cells |
CD45+ CD3- CD19+ CD20+ HLA-DR+ CD27+ |
|
Early NK |
CD45+ CD3- CD19- CD14- CD56+ CD16- |
|
Mature NK |
CD45+ CD3- CD19- CD14- CD56+ CD16+ |
|
M-MDSCs |
CD45+ CD3- CD19- CD11b+ CD33+ CD14+ HLA-DR-/dim |
|
Classical Monos |
CD45+ CD3- CD19- CD56- CD11b+ CD33+ HLA-DR+ CD14+ CD16- |
|
Intermediate Monos |
CD45+ CD3- CD19- CD56- CD11b+ CD33+ HLA-DR+ CD14+ CD16+ |
|
Non-Classical Monos |
CD45+ CD3- CD19- CD56- CD11b+ CD33+ HLA-DR+ CD14- /dim CD16++ |
|
RV in Monos |
CD45+ CD3- CD19- CD56- CD11b+ CD33+ HLA-DR+ CD14+ RV+ |
|
NKt-cells |
CD45+ CD19- CD3+ CD56+ |
|
TCR γδ |
CD45+ CD19- CD3+ CD56- TCRgd+ |
|
AE T-cells |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25+ CD127+ |
|
Memory Tregs |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25+ CD127low/- CD45RA- |
|
Naïve Tregs |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25+ CD127low/- CD45RA+ |
|
CM CD8 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8+ CD4- CD45RA- CCR7+ CD27+ |
|
EM CD8 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8+ CD4- CD45RA- CCR7- CD27+ |
|
Naïve CD8 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8+ CD4- CD45RA+ CCR7+ CD27+ |
|
TEMRA CD8 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8+ CD4- CD45RA+ CCR7- CD27- |
|
CM CD4 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25- CD127+CD45RA- CCR7+ CD27+ |
|
EM CD4 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25- CD127+CD45RA- CCR7- CD27+ |
|
Naïve CD4 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25- CD127+CD45RA+ CCR7+ CD27+ |
|
TEMRA CD4 |
CD45+ CD19- CD3+ CD56- TCRgd- CD8- CD4+ CD25- CD127+/dim CD45RA+ CCR7- CD27- |
In both patients, we observed a substantial decrease in the total number of B cells following leflunomide treatment (Fig. 2B and Sup. Fig. 2D) (Sup. Tables 3 and 4). This finding is consistent with the regulatory activity of leflunomide on B cell proliferation that was previously observed in animals11. Although leflunomide has been considered an immunosuppressive drug by inhibiting cellular and humoral mediated responses12, in both patients, we observed a robust expansion in CD8+ cytotoxic T cells after 5 days from the initial dosing. This effect further increased upon subsequent doses, reaching maximum expansion at around 21 days after the initial treatment (Fig. 2C and Sup. Fig. 2E-G). Among the CD8+ cytotoxic T cells, the major increase was observed in the effector memory (EM) and in the CD45RA+ terminal effector T cell population (TEMRA CD8+) (Fig. 2D), whereas the other T cell populations, including naïve and central memory (CM), were nearly unaffected. These data are wholly aligned with recently published data13 showing that expansion of CD8+ cytotoxic T cells, including long lived, antigen-experienced T cells (CD45RA+), contribute to SARS-CoV-2 recovery in cancer patients. Intriguingly, in three patients with relapsed multiple myeloma enrolled in our single agent leflunomide phase 1 trial9, we observed that leflunomide elicited the same immune stimulatory effect on CD8+ cytotoxic T cells (unpublished data), further supporting an unexpected immune stimulatory role of this agent in cancer patients.
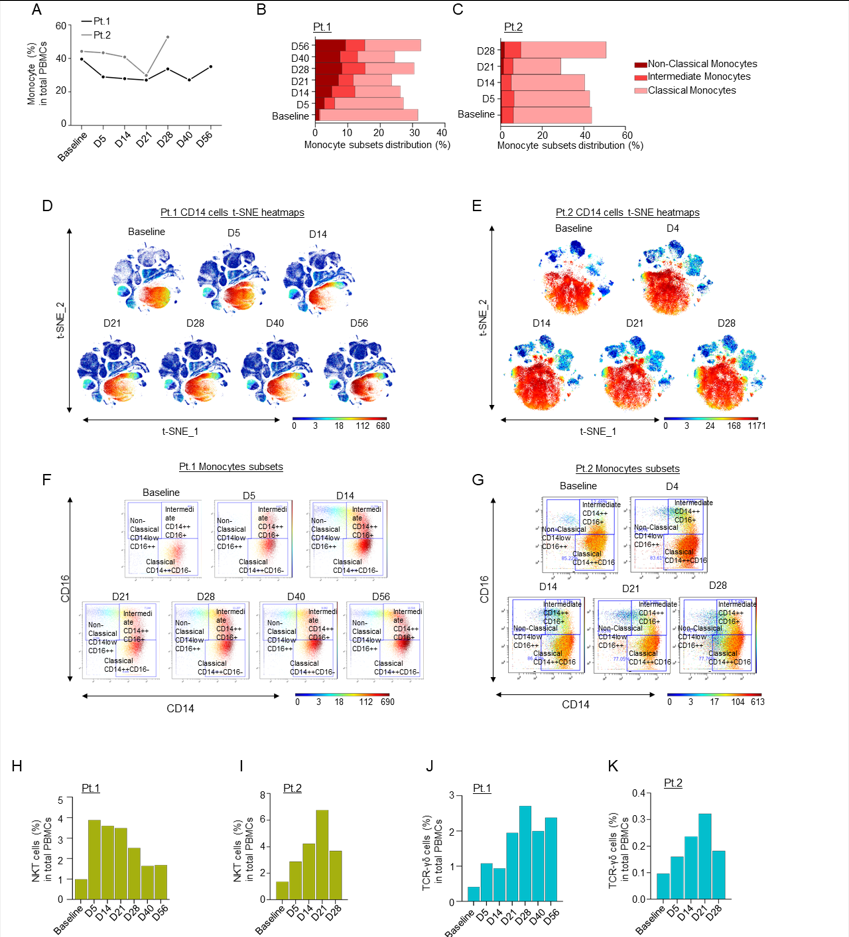
Supplementary Figure 3: A) Line graph showing percentages of monocyte cells in PBMCs in both patients; B-C) Bar graphs showing percentages of monocyte cell immune sub-compartments, based on CD14 and CD16 expression; D-E) t-SNE heatmaps showing overall CD14 expression through the course of treatment in patients 1 and 2; F-G) Dot plots colored by CD14 channel for the signal of Classical, Intermediate and Non-Classical monocyte expression through the course of treatment. H-I) Bar graphs showing percentages of NKT cells compared to the total count PBMCs in Patients 1 (H) and 2 (I) over time, as measured by CyTOF; J-K) Bar graphs showing percentages of TCR-γδ cells compared to the total count PBMC in Patients 1 (J) and 2 (K) over time, as measured by CyTOF.
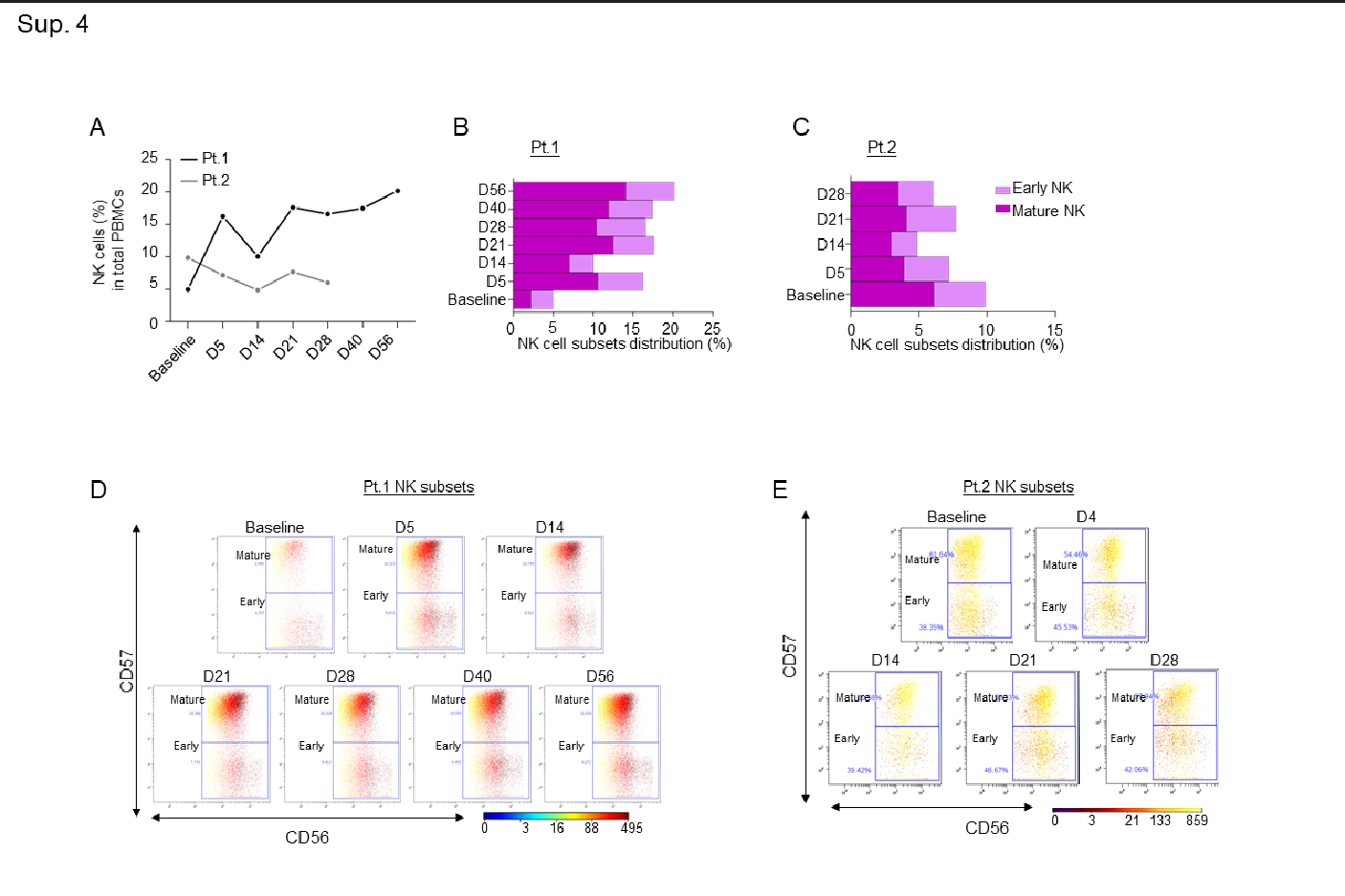
Supplementary Figure 4: A-B-C) Line graph showing percentages of NK cells in both patients (A), and bar graphs showing percentages of early and mature NK cell immune sub-compartments in both patients at the indicated timepoints (B-C); D-E) Dot plots colored by CD56 channel for the signal of early and mature NK cell expression through the course of treatment.
Supplementary Table 3: CyTOF counts, Patient 1
|
Timepoint |
Population |
FCS Filename |
Event count |
|
Baseline |
Leukocytes |
20291_PB_Basline |
151061 |
|
Baseline |
Granulocytes |
20291_PB_Basline |
0 |
|
Baseline |
PBMCs |
20291_PB_Basline |
151061 |
|
Baseline |
B-cells |
20291_PB_Basline |
6986 |
|
Baseline |
Naive B-cells |
20291_PB_Basline |
4515 |
|
Baseline |
Total Memory B-cells |
20291_PB_Basline |
2471 |
|
Baseline |
Memory B-cells |
20291_PB_Basline |
246 |
|
Baseline |
Plasmablast |
20291_PB_Basline |
2160 |
|
Baseline |
CD3- CD19- |
20291_PB_Basline |
85407 |
|
Baseline |
CD56+ CD14+ |
20291_PB_Basline |
18060 |
|
Baseline |
NK-cells |
20291_PB_Basline |
7512 |
|
Baseline |
Early NK |
20291_PB_Basline |
4185 |
|
Baseline |
Mature NK |
20291_PB_Basline |
3322 |
|
Baseline |
Monocytes ? |
20291_PB_Basline |
59803 |
|
Baseline |
Classical Monocytes |
20291_PB_Basline |
45571 |
|
Baseline |
Intermediate Monocytes |
20291_PB_Basline |
412 |
|
Baseline |
Non-Classical Monocytes |
20291_PB_Basline |
1858 |
|
Baseline |
CD14- CD16- |
20291_PB_Basline |
11712 |
|
Baseline |
Myeloid Dendritic cells |
20291_PB_Basline |
1604 |
|
Baseline |
Plasmacytoid Dendritic cells |
20291_PB_Basline |
47 |
|
Baseline |
CD3+ |
20291_PB_Basline |
58560 |
|
Baseline |
NKT |
20291_PB_Basline |
1486 |
|
Baseline |
T-cells |
20291_PB_Basline |
57072 |
|
Baseline |
TCR g/d |
20291_PB_Basline |
620 |
|
Baseline |
TCR a/b |
20291_PB_Basline |
56452 |
|
Baseline |
CD8 T-cells |
20291_PB_Basline |
19802 |
|
Baseline |
Effector CD8 T-cells |
20291_PB_Basline |
1220 |
|
Baseline |
Naive CD8 |
20291_PB_Basline |
7996 |
|
Baseline |
CD8+ CD27- |
20291_PB_Basline |
5143 |
|
Baseline |
Late differentiated CD8 T-cells |
20291_PB_Basline |
1650 |
|
Baseline |
Temra CD8 |
20291_PB_Basline |
3488 |
|
Baseline |
CD8+ CD45RA- CD27+ |
20291_PB_Basline |
5450 |
|
Baseline |
CM CD8 |
20291_PB_Basline |
2317 |
|
Baseline |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_Basline |
3123 |
|
Baseline |
EM CD8 |
20291_PB_Basline |
1253 |
|
Baseline |
Transitional memory CD8 T-cells |
20291_PB_Basline |
1870 |
|
Baseline |
CD4 T-cells |
20291_PB_Basline |
35741 |
|
Baseline |
CD4+ CD25+ |
20291_PB_Basline |
1649 |
|
Baseline |
Activated Effector CD4 T-cells |
20291_PB_Basline |
319 |
|
Baseline |
Total Tregs |
20291_PB_Basline |
1330 |
|
Baseline |
Memory Tregs |
20291_PB_Basline |
1301 |
|
Baseline |
Naive Tregs |
20291_PB_Basline |
29 |
|
Baseline |
CD4+ CD25- |
20291_PB_Basline |
34092 |
|
Baseline |
CM CD4 T-cells |
20291_PB_Basline |
16059 |
|
Baseline |
Naive CD4 T-cells |
20291_PB_Basline |
10541 |
|
Baseline |
CD4+ CCR7- CD45RA? |
20291_PB_Basline |
7473 |
|
Baseline |
EM CD4 |
20291_PB_Basline |
5353 |
|
Baseline |
Temra CD4 |
20291_PB_Basline |
2120 |
|
D5 |
Leukocytes |
20291_PB_D5 |
151061 |
|
D5 |
Granulocytes |
20291_PB_D5 |
0 |
|
D5 |
PBMCs |
20291_PB_D5 |
151061 |
|
D5 |
B-cells |
20291_PB_D5 |
2667 |
|
D5 |
Naive B-cells |
20291_PB_D5 |
1766 |
|
D5 |
Total Memory B-cells |
20291_PB_D5 |
901 |
|
D5 |
Memory B-cells |
20291_PB_D5 |
250 |
|
D5 |
Plasmablast |
20291_PB_D5 |
605 |
|
D5 |
CD3- CD19- |
20291_PB_D5 |
80611 |
|
D5 |
CD56+ CD14+ |
20291_PB_D5 |
12229 |
|
D5 |
NK-cells |
20291_PB_D5 |
24543 |
|
D5 |
Early NK |
20291_PB_D5 |
8525 |
|
D5 |
Mature NK |
20291_PB_D5 |
15992 |
|
D5 |
Monocytes ? |
20291_PB_D5 |
43783 |
|
D5 |
Classical Monocytes |
20291_PB_D5 |
32035 |
|
D5 |
Intermediate Monocytes |
20291_PB_D5 |
4817 |
|
D5 |
Non-Classical Monocytes |
20291_PB_D5 |
4244 |
|
D5 |
CD14- CD16- |
20291_PB_D5 |
2628 |
|
D5 |
Myeloid Dendritic cells |
20291_PB_D5 |
547 |
|
D5 |
Plasmacytoid Dendritic cells |
20291_PB_D5 |
31 |
|
D5 |
CD3+ |
20291_PB_D5 |
67731 |
|
D5 |
NKT |
20291_PB_D5 |
5856 |
|
D5 |
T-cells |
20291_PB_D5 |
61874 |
|
D5 |
TCR g/d |
20291_PB_D5 |
1629 |
|
D5 |
TCR a/b |
20291_PB_D5 |
60245 |
|
D5 |
CD8 T-cells |
20291_PB_D5 |
36652 |
|
D5 |
Effector CD8 T-cells |
20291_PB_D5 |
4553 |
|
D5 |
Naive CD8 |
20291_PB_D5 |
5435 |
|
D5 |
CD8+ CD27- |
20291_PB_D5 |
21065 |
|
D5 |
Late differentiated CD8 T-cells |
20291_PB_D5 |
5815 |
|
D5 |
Temra CD8 |
20291_PB_D5 |
15243 |
|
D5 |
CD8+ CD45RA- CD27+ |
20291_PB_D5 |
5613 |
|
D5 |
CM CD8 |
20291_PB_D5 |
1273 |
|
D5 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D5 |
4334 |
|
D5 |
EM CD8 |
20291_PB_D5 |
2579 |
|
D5 |
Transitional memory CD8 T-cells |
20291_PB_D5 |
1755 |
|
D5 |
CD4 T-cells |
20291_PB_D5 |
22510 |
|
D5 |
CD4+ CD25+ |
20291_PB_D5 |
856 |
|
D5 |
Activated Effector CD4 T-cells |
20291_PB_D5 |
228 |
|
D5 |
Total Tregs |
20291_PB_D5 |
628 |
|
D5 |
Memory Tregs |
20291_PB_D5 |
617 |
|
D5 |
Naive Tregs |
20291_PB_D5 |
11 |
|
D5 |
CD4+ CD25- |
20291_PB_D5 |
21654 |
|
D5 |
CM CD4 T-cells |
20291_PB_D5 |
8378 |
|
D5 |
Naive CD4 T-cells |
20291_PB_D5 |
5390 |
|
D5 |
CD4+ CCR7- CD45RA? |
20291_PB_D5 |
7874 |
|
D5 |
EM CD4 |
20291_PB_D5 |
4788 |
|
D5 |
Temra CD4 |
20291_PB_D5 |
3086 |
|
D8 |
Leukocytes |
20291_PB_D8 |
151061 |
|
D8 |
Granulocytes |
20291_PB_D8 |
0 |
|
D8 |
PBMCs |
20291_PB_D8 |
151061 |
|
D8 |
B-cells |
20291_PB_D8 |
2670 |
|
D8 |
Naive B-cells |
20291_PB_D8 |
2093 |
|
D8 |
Total Memory B-cells |
20291_PB_D8 |
577 |
|
D8 |
Memory B-cells |
20291_PB_D8 |
384 |
|
D8 |
Plasmablast |
20291_PB_D8 |
154 |
|
D8 |
CD3- CD19- |
20291_PB_D8 |
76613 |
|
D8 |
CD56+ CD14+ |
20291_PB_D8 |
14437 |
|
D8 |
NK-cells |
20291_PB_D8 |
12448 |
|
D8 |
Early NK |
20291_PB_D8 |
4902 |
|
D8 |
Mature NK |
20291_PB_D8 |
7536 |
|
D8 |
Monocytes ? |
20291_PB_D8 |
49677 |
|
D8 |
Classical Monocytes |
20291_PB_D8 |
40484 |
|
D8 |
Intermediate Monocytes |
20291_PB_D8 |
1687 |
|
D8 |
Non-Classical Monocytes |
20291_PB_D8 |
2666 |
|
D8 |
CD14- CD16- |
20291_PB_D8 |
4737 |
|
D8 |
Myeloid Dendritic cells |
20291_PB_D8 |
576 |
|
D8 |
Plasmacytoid Dendritic cells |
20291_PB_D8 |
75 |
|
D8 |
CD3+ |
20291_PB_D8 |
71737 |
|
D8 |
NKT |
20291_PB_D8 |
4110 |
|
D8 |
T-cells |
20291_PB_D8 |
67625 |
|
D8 |
TCR g/d |
20291_PB_D8 |
1291 |
|
D8 |
TCR a/b |
20291_PB_D8 |
66334 |
|
D8 |
CD8 T-cells |
20291_PB_D8 |
30916 |
|
D8 |
Effector CD8 T-cells |
20291_PB_D8 |
3711 |
|
D8 |
Naive CD8 |
20291_PB_D8 |
6102 |
|
D8 |
CD8+ CD27- |
20291_PB_D8 |
12227 |
|
D8 |
Late differentiated CD8 T-cells |
20291_PB_D8 |
3473 |
|
D8 |
Temra CD8 |
20291_PB_D8 |
8754 |
|
D8 |
CD8+ CD45RA- CD27+ |
20291_PB_D8 |
8882 |
|
D8 |
CM CD8 |
20291_PB_D8 |
3057 |
|
D8 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D8 |
5814 |
|
D8 |
EM CD8 |
20291_PB_D8 |
2298 |
|
D8 |
Transitional memory CD8 T-cells |
20291_PB_D8 |
3516 |
|
D8 |
CD4 T-cells |
20291_PB_D8 |
34085 |
|
D8 |
CD4+ CD25+ |
20291_PB_D8 |
1257 |
|
D8 |
Activated Effector CD4 T-cells |
20291_PB_D8 |
285 |
|
D8 |
Total Tregs |
20291_PB_D8 |
972 |
|
D8 |
Memory Tregs |
20291_PB_D8 |
958 |
|
D8 |
Naive Tregs |
20291_PB_D8 |
14 |
|
D8 |
CD4+ CD25- |
20291_PB_D8 |
32828 |
|
D8 |
CM CD4 T-cells |
20291_PB_D8 |
15459 |
|
D8 |
Naive CD4 T-cells |
20291_PB_D8 |
5126 |
|
D8 |
CD4+ CCR7- CD45RA? |
20291_PB_D8 |
12220 |
|
D8 |
EM CD4 |
20291_PB_D8 |
8907 |
|
D8 |
Temra CD4 |
20291_PB_D8 |
3313 |
|
D14 |
Leukocytes |
20291_PB_D14 |
151061 |
|
D14 |
Granulocytes |
20291_PB_D14 |
0 |
|
D14 |
PBMCs |
20291_PB_D14 |
151061 |
|
D14 |
B-cells |
20291_PB_D14 |
897 |
|
D14 |
Naive B-cells |
20291_PB_D14 |
765 |
|
D14 |
Total Memory B-cells |
20291_PB_D14 |
132 |
|
D14 |
Memory B-cells |
20291_PB_D14 |
104 |
|
D14 |
Plasmablast |
20291_PB_D14 |
26 |
|
D14 |
CD3- CD19- |
20291_PB_D14 |
58555 |
|
D14 |
CD56+ CD14+ |
20291_PB_D14 |
1185 |
|
D14 |
NK-cells |
20291_PB_D14 |
15179 |
|
D14 |
Early NK |
20291_PB_D14 |
4511 |
|
D14 |
Mature NK |
20291_PB_D14 |
10650 |
|
D14 |
Monocytes ? |
20291_PB_D14 |
42158 |
|
D14 |
Classical Monocytes |
20291_PB_D14 |
21098 |
|
D14 |
Intermediate Monocytes |
20291_PB_D14 |
10818 |
|
D14 |
Non-Classical Monocytes |
20291_PB_D14 |
7742 |
|
D14 |
CD14- CD16- |
20291_PB_D14 |
2534 |
|
D14 |
Myeloid Dendritic cells |
20291_PB_D14 |
1199 |
|
D14 |
Plasmacytoid Dendritic cells |
20291_PB_D14 |
800 |
|
D14 |
CD3+ |
20291_PB_D14 |
91559 |
|
D14 |
NKT |
20291_PB_D14 |
5436 |
|
D14 |
T-cells |
20291_PB_D14 |
86108 |
|
D14 |
TCR g/d |
20291_PB_D14 |
1418 |
|
D14 |
TCR a/b |
20291_PB_D14 |
84690 |
|
D14 |
CD8 T-cells |
20291_PB_D14 |
46721 |
|
D14 |
Effector CD8 T-cells |
20291_PB_D14 |
6100 |
|
D14 |
Naive CD8 |
20291_PB_D14 |
8287 |
|
D14 |
CD8+ CD27- |
20291_PB_D14 |
17649 |
|
D14 |
Late differentiated CD8 T-cells |
20291_PB_D14 |
4910 |
|
D14 |
Temra CD8 |
20291_PB_D14 |
12739 |
|
D14 |
CD8+ CD45RA- CD27+ |
20291_PB_D14 |
14706 |
|
D14 |
CM CD8 |
20291_PB_D14 |
3339 |
|
D14 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D14 |
11354 |
|
D14 |
EM CD8 |
20291_PB_D14 |
5743 |
|
D14 |
Transitional memory CD8 T-cells |
20291_PB_D14 |
5611 |
|
D14 |
CD4 T-cells |
20291_PB_D14 |
36245 |
|
D14 |
CD4+ CD25+ |
20291_PB_D14 |
1234 |
|
D14 |
Activated Effector CD4 T-cells |
20291_PB_D14 |
165 |
|
D14 |
Total Tregs |
20291_PB_D14 |
1069 |
|
D14 |
Memory Tregs |
20291_PB_D14 |
939 |
|
D14 |
Naive Tregs |
20291_PB_D14 |
130 |
|
D14 |
CD4+ CD25- |
20291_PB_D14 |
35011 |
|
D14 |
CM CD4 T-cells |
20291_PB_D14 |
16288 |
|
D14 |
Naive CD4 T-cells |
20291_PB_D14 |
10797 |
|
D14 |
CD4+ CCR7- CD45RA? |
20291_PB_D14 |
7899 |
|
D14 |
EM CD4 |
20291_PB_D14 |
4888 |
|
D14 |
Temra CD4 |
20291_PB_D14 |
3011 |
|
D21 |
Leukocytes |
20291_PB_D21 |
151061 |
|
D21 |
Granulocytes |
20291_PB_D21 |
0 |
|
D21 |
PBMCs |
20291_PB_D21 |
151061 |
|
D21 |
B-cells |
20291_PB_D21 |
607 |
|
D21 |
Naive B-cells |
20291_PB_D21 |
564 |
|
D21 |
Total Memory B-cells |
20291_PB_D21 |
43 |
|
D21 |
Memory B-cells |
20291_PB_D21 |
37 |
|
D21 |
Plasmablast |
20291_PB_D21 |
4 |
|
D21 |
CD3- CD19- |
20291_PB_D21 |
68944 |
|
D21 |
CD56+ CD14+ |
20291_PB_D21 |
1396 |
|
D21 |
NK-cells |
20291_PB_D21 |
26605 |
|
D21 |
Early NK |
20291_PB_D21 |
7638 |
|
D21 |
Mature NK |
20291_PB_D21 |
18946 |
|
D21 |
Monocytes ? |
20291_PB_D21 |
40864 |
|
D21 |
Classical Monocytes |
20291_PB_D21 |
17955 |
|
D21 |
Intermediate Monocytes |
20291_PB_D21 |
6880 |
|
D21 |
Non-Classical Monocytes |
20291_PB_D21 |
10871 |
|
D21 |
CD14- CD16- |
20291_PB_D21 |
5147 |
|
D21 |
Myeloid Dendritic cells |
20291_PB_D21 |
1701 |
|
D21 |
Plasmacytoid Dendritic cells |
20291_PB_D21 |
1644 |
|
D21 |
CD3+ |
20291_PB_D21 |
81442 |
|
D21 |
NKT |
20291_PB_D21 |
5272 |
|
D21 |
T-cells |
20291_PB_D21 |
76163 |
|
D21 |
TCR g/d |
20291_PB_D21 |
2945 |
|
D21 |
TCR a/b |
20291_PB_D21 |
73218 |
|
D21 |
CD8 T-cells |
20291_PB_D21 |
45223 |
|
D21 |
Effector CD8 T-cells |
20291_PB_D21 |
10028 |
|
D21 |
Naive CD8 |
20291_PB_D21 |
5742 |
|
D21 |
CD8+ CD27- |
20291_PB_D21 |
18462 |
|
D21 |
Late differentiated CD8 T-cells |
20291_PB_D21 |
5045 |
|
D21 |
Temra CD8 |
20291_PB_D21 |
13414 |
|
D21 |
CD8+ CD45RA- CD27+ |
20291_PB_D21 |
11026 |
|
D21 |
CM CD8 |
20291_PB_D21 |
1706 |
|
D21 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D21 |
9310 |
|
D21 |
EM CD8 |
20291_PB_D21 |
6616 |
|
D21 |
Transitional memory CD8 T-cells |
20291_PB_D21 |
2694 |
|
D21 |
CD4 T-cells |
20291_PB_D21 |
26209 |
|
D21 |
CD4+ CD25+ |
20291_PB_D21 |
1477 |
|
D21 |
Activated Effector CD4 T-cells |
20291_PB_D21 |
206 |
|
D21 |
Total Tregs |
20291_PB_D21 |
1271 |
|
D21 |
Memory Tregs |
20291_PB_D21 |
1168 |
|
D21 |
Naive Tregs |
20291_PB_D21 |
103 |
|
D21 |
CD4+ CD25- |
20291_PB_D21 |
24732 |
|
D21 |
CM CD4 T-cells |
20291_PB_D21 |
8595 |
|
D21 |
Naive CD4 T-cells |
20291_PB_D21 |
6552 |
|
D21 |
CD4+ CCR7- CD45RA? |
20291_PB_D21 |
9574 |
|
D21 |
EM CD4 |
20291_PB_D21 |
3545 |
|
D21 |
Temra CD4 |
20291_PB_D21 |
6029 |
|
D28 |
Leukocytes |
20291_PB_D28 |
151061 |
|
D28 |
Granulocytes |
20291_PB_D28 |
0 |
|
D28 |
PBMCs |
20291_PB_D28 |
151061 |
|
D28 |
B-cells |
20291_PB_D28 |
476 |
|
D28 |
Naive B-cells |
20291_PB_D28 |
448 |
|
D28 |
Total Memory B-cells |
20291_PB_D28 |
28 |
|
D28 |
Memory B-cells |
20291_PB_D28 |
21 |
|
D28 |
Plasmablast |
20291_PB_D28 |
7 |
|
D28 |
CD3- CD19- |
20291_PB_D28 |
77699 |
|
D28 |
CD56+ CD14+ |
20291_PB_D28 |
1618 |
|
D28 |
NK-cells |
20291_PB_D28 |
25055 |
|
D28 |
Early NK |
20291_PB_D28 |
9258 |
|
D28 |
Mature NK |
20291_PB_D28 |
15773 |
|
D28 |
Monocytes ? |
20291_PB_D28 |
50952 |
|
D28 |
Classical Monocytes |
20291_PB_D28 |
22859 |
|
D28 |
Intermediate Monocytes |
20291_PB_D28 |
10833 |
|
D28 |
Non-Classical Monocytes |
20291_PB_D28 |
12452 |
|
D28 |
CD14- CD16- |
20291_PB_D28 |
4828 |
|
D28 |
Myeloid Dendritic cells |
20291_PB_D28 |
1566 |
|
D28 |
Plasmacytoid Dendritic cells |
20291_PB_D28 |
1749 |
|
D28 |
CD3+ |
20291_PB_D28 |
72834 |
|
D28 |
NKT |
20291_PB_D28 |
3806 |
|
D28 |
T-cells |
20291_PB_D28 |
69023 |
|
D28 |
TCR g/d |
20291_PB_D28 |
4092 |
|
D28 |
TCR a/b |
20291_PB_D28 |
64931 |
|
D28 |
CD8 T-cells |
20291_PB_D28 |
39848 |
|
D28 |
Effector CD8 T-cells |
20291_PB_D28 |
12908 |
|
D28 |
Naive CD8 |
20291_PB_D28 |
4526 |
|
D28 |
CD8+ CD27- |
20291_PB_D28 |
14265 |
|
D28 |
Late differentiated CD8 T-cells |
20291_PB_D28 |
4261 |
|
D28 |
Temra CD8 |
20291_PB_D28 |
10004 |
|
D28 |
CD8+ CD45RA- CD27+ |
20291_PB_D28 |
8160 |
|
D28 |
CM CD8 |
20291_PB_D28 |
1624 |
|
D28 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D28 |
6524 |
|
D28 |
EM CD8 |
20291_PB_D28 |
4572 |
|
D28 |
Transitional memory CD8 T-cells |
20291_PB_D28 |
1952 |
|
D28 |
CD4 T-cells |
20291_PB_D28 |
23246 |
|
D28 |
CD4+ CD25+ |
20291_PB_D28 |
1010 |
|
D28 |
Activated Effector CD4 T-cells |
20291_PB_D28 |
128 |
|
D28 |
Total Tregs |
20291_PB_D28 |
882 |
|
D28 |
Memory Tregs |
20291_PB_D28 |
835 |
|
D28 |
Naive Tregs |
20291_PB_D28 |
47 |
|
D28 |
CD4+ CD25- |
20291_PB_D28 |
22236 |
|
D28 |
CM CD4 T-cells |
20291_PB_D28 |
8490 |
|
D28 |
Naive CD4 T-cells |
20291_PB_D28 |
5365 |
|
D28 |
CD4+ CCR7- CD45RA? |
20291_PB_D28 |
8371 |
|
D28 |
EM CD4 |
20291_PB_D28 |
3362 |
|
D28 |
Temra CD4 |
20291_PB_D28 |
5009 |
|
D40 |
Leukocytes |
20291_PB_D40 |
151061 |
|
D40 |
Granulocytes |
20291_PB_D40 |
0 |
|
D40 |
PBMCs |
20291_PB_D40 |
151061 |
|
D40 |
B-cells |
20291_PB_D40 |
689 |
|
D40 |
Naive B-cells |
20291_PB_D40 |
425 |
|
D40 |
Total Memory B-cells |
20291_PB_D40 |
264 |
|
D40 |
Memory B-cells |
20291_PB_D40 |
38 |
|
D40 |
Plasmablast |
20291_PB_D40 |
206 |
|
D40 |
CD3- CD19- |
20291_PB_D40 |
69036 |
|
D40 |
CD56+ CD14+ |
20291_PB_D40 |
1481 |
|
D40 |
NK-cells |
20291_PB_D40 |
26394 |
|
D40 |
Early NK |
20291_PB_D40 |
8310 |
|
D40 |
Mature NK |
20291_PB_D40 |
18050 |
|
D40 |
Monocytes ? |
20291_PB_D40 |
41087 |
|
D40 |
Classical Monocytes |
20291_PB_D40 |
17194 |
|
D40 |
Intermediate Monocytes |
20291_PB_D40 |
8146 |
|
D40 |
Non-Classical Monocytes |
20291_PB_D40 |
11709 |
|
D40 |
CD14- CD16- |
20291_PB_D40 |
4072 |
|
D40 |
Myeloid Dendritic cells |
20291_PB_D40 |
1331 |
|
D40 |
Plasmacytoid Dendritic cells |
20291_PB_D40 |
644 |
|
D40 |
CD3+ |
20291_PB_D40 |
81280 |
|
D40 |
NKT |
20291_PB_D40 |
2476 |
|
D40 |
T-cells |
20291_PB_D40 |
78802 |
|
D40 |
TCR g/d |
20291_PB_D40 |
3016 |
|
D40 |
TCR a/b |
20291_PB_D40 |
75786 |
|
D40 |
CD8 T-cells |
20291_PB_D40 |
40159 |
|
D40 |
Effector CD8 T-cells |
20291_PB_D40 |
10555 |
|
D40 |
Naive CD8 |
20291_PB_D40 |
6122 |
|
D40 |
CD8+ CD27- |
20291_PB_D40 |
14136 |
|
D40 |
Late differentiated CD8 T-cells |
20291_PB_D40 |
3103 |
|
D40 |
Temra CD8 |
20291_PB_D40 |
11015 |
|
D40 |
CD8+ CD45RA- CD27+ |
20291_PB_D40 |
9392 |
|
D40 |
CM CD8 |
20291_PB_D40 |
2300 |
|
D40 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D40 |
7082 |
|
D40 |
EM CD8 |
20291_PB_D40 |
4816 |
|
D40 |
Transitional memory CD8 T-cells |
20291_PB_D40 |
2266 |
|
D40 |
CD4 T-cells |
20291_PB_D40 |
33775 |
|
D40 |
CD4+ CD25+ |
20291_PB_D40 |
952 |
|
D40 |
Activated Effector CD4 T-cells |
20291_PB_D40 |
134 |
|
D40 |
Total Tregs |
20291_PB_D40 |
818 |
|
D40 |
Memory Tregs |
20291_PB_D40 |
749 |
|
D40 |
Naive Tregs |
20291_PB_D40 |
69 |
|
D40 |
CD4+ CD25- |
20291_PB_D40 |
32823 |
|
D40 |
CM CD4 T-cells |
20291_PB_D40 |
12546 |
|
D40 |
Naive CD4 T-cells |
20291_PB_D40 |
7278 |
|
D40 |
CD4+ CCR7- CD45RA? |
20291_PB_D40 |
12963 |
|
D40 |
EM CD4 |
20291_PB_D40 |
7191 |
|
D40 |
Temra CD4 |
20291_PB_D40 |
5772 |
|
D56 |
Leukocytes |
20291_PB_D56 |
151061 |
|
D56 |
Granulocytes |
20291_PB_D56 |
0 |
|
D56 |
PBMCs |
20291_PB_D56 |
151061 |
|
D56 |
B-cells |
20291_PB_D56 |
1205 |
|
D56 |
Naive B-cells |
20291_PB_D56 |
805 |
|
D56 |
Total Memory B-cells |
20291_PB_D56 |
400 |
|
D56 |
Memory B-cells |
20291_PB_D56 |
66 |
|
D56 |
Plasmablast |
20291_PB_D56 |
292 |
|
D56 |
CD3- CD19- |
20291_PB_D56 |
85540 |
|
D56 |
CD56+ CD14+ |
20291_PB_D56 |
1927 |
|
D56 |
NK-cells |
20291_PB_D56 |
30493 |
|
D56 |
Early NK |
20291_PB_D56 |
9146 |
|
D56 |
Mature NK |
20291_PB_D56 |
21311 |
|
D56 |
Monocytes ? |
20291_PB_D56 |
53028 |
|
D56 |
Classical Monocytes |
20291_PB_D56 |
26053 |
|
D56 |
Intermediate Monocytes |
20291_PB_D56 |
8935 |
|
D56 |
Non-Classical Monocytes |
20291_PB_D56 |
14216 |
|
D56 |
CD14- CD16- |
20291_PB_D56 |
3861 |
|
D56 |
Myeloid Dendritic cells |
20291_PB_D56 |
1022 |
|
D56 |
Plasmacytoid Dendritic cells |
20291_PB_D56 |
1979 |
|
D56 |
CD3+ |
20291_PB_D56 |
64265 |
|
D56 |
NKT |
20291_PB_D56 |
2541 |
|
D56 |
T-cells |
20291_PB_D56 |
61720 |
|
D56 |
TCR g/d |
20291_PB_D56 |
3596 |
|
D56 |
TCR a/b |
20291_PB_D56 |
58124 |
|
D56 |
CD8 T-cells |
20291_PB_D56 |
30821 |
|
D56 |
Effector CD8 T-cells |
20291_PB_D56 |
7494 |
|
D56 |
Naive CD8 |
20291_PB_D56 |
7316 |
|
D56 |
CD8+ CD27- |
20291_PB_D56 |
10681 |
|
D56 |
Late differentiated CD8 T-cells |
20291_PB_D56 |
2670 |
|
D56 |
Temra CD8 |
20291_PB_D56 |
8011 |
|
D56 |
CD8+ CD45RA- CD27+ |
20291_PB_D56 |
5350 |
|
D56 |
CM CD8 |
20291_PB_D56 |
1911 |
|
D56 |
CD8+ CD45RA- CD27+ CCR7- |
20291_PB_D56 |
3430 |
|
D56 |
EM CD8 |
20291_PB_D56 |
2230 |
|
D56 |
Transitional memory CD8 T-cells |
20291_PB_D56 |
1200 |
|
D56 |
CD4 T-cells |
20291_PB_D56 |
25412 |
|
D56 |
CD4+ CD25+ |
20291_PB_D56 |
1293 |
|
D56 |
Activated Effector CD4 T-cells |
20291_PB_D56 |
114 |
|
D56 |
Total Tregs |
20291_PB_D56 |
1179 |
|
D56 |
Memory Tregs |
20291_PB_D56 |
1084 |
|
D56 |
Naive Tregs |
20291_PB_D56 |
95 |
|
D56 |
CD4+ CD25- |
20291_PB_D56 |
24119 |
|
D56 |
CM CD4 T-cells |
20291_PB_D56 |
8984 |
|
D56 |
Naive CD4 T-cells |
20291_PB_D56 |
8027 |
|
D56 |
CD4+ CCR7- CD45RA? |
20291_PB_D56 |
7095 |
|
D56 |
EM CD4 |
20291_PB_D56 |
3532 |
|
D56 |
Temra CD4 |
20291_PB_D56 |
3563 |
Supplementary Table 4: CyTOF Counts, Patient 2
|
Population |
Timepoint |
FCS Filename |
Event count |
|
PBMCs |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
95678 |
|
PBMCs |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
95678 |
|
PBMCs |
D8 |
Pt2_D8_PBMC_merge1 |
95678 |
|
PBMCs |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
95678 |
|
PBMCs |
D21 |
Pt.#2_D21_PBMC_IRB20291_MDIPA_10-13-2021_1 |
95678 |
|
PBMCs |
D28 |
Pt.#2_D28_PBMC_IRB20291_MDIPA_10-13-2021_1 |
95678 |
|
B-cells |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
12212 |
|
B-cells |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
5966 |
|
B-cells |
D8 |
Pt2_D8_PBMC_merge1 |
3561 |
|
B-cells |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
4293 |
|
B-cells |
D21 |
Pt.#2_D21_PBMC_IRB20291_MDIPA_10-13-2021_1 |
5977 |
|
B-cells |
D28 |
Pt.#2_D28_PBMC_IRB20291_MDIPA_10-13-2021_1 |
2283 |
|
Naive B-cells |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
12020 |
|
Naive B-cells |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
5661 |
|
Naive B-cells |
D8 |
Pt2_D8_PBMC_merge1 |
3528 |
|
Naive B-cells |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
3865 |
|
Naive B-cells |
D21 |
Pt.#2_D21_PBMC_IRB20291_MDIPA_10-13-2021_1 |
5140 |
|
Naive B-cells |
D28 |
Pt.#2_D28_PBMC_IRB20291_MDIPA_10-13-2021_1 |
2232 |
|
Total Memory B-cells |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
191 |
|
Total Memory B-cells |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
305 |
|
Total Memory B-cells |
D8 |
Pt2_D8_PBMC_merge1 |
32 |
|
Total Memory B-cells |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
427 |
|
Total Memory B-cells |
D21 |
Pt.#2_D21_PBMC_IRB20291_MDIPA_10-13-2021_1 |
835 |
|
Total Memory B-cells |
D28 |
Pt.#2_D28_PBMC_IRB20291_MDIPA_10-13-2021_1 |
51 |
|
Plasmablast |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
47 |
|
Plasmablast |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
5 |
|
Plasmablast |
D8 |
Pt2_D8_PBMC_merge1 |
4 |
|
Plasmablast |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
8 |
|
Plasmablast |
D21 |
Pt.#2_D21_PBMC_IRB20291_MDIPA_10-13-2021_1 |
23 |
|
Plasmablast |
D28 |
Pt.#2_D28_PBMC_IRB20291_MDIPA_10-13-2021_1 |
7 |
|
NK-cells |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
9448 |
|
NK-cells |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
6875 |
|
NK-cells |
D8 |
Pt2_D8_PBMC_merge1 |
11434 |
|
NK-cells |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
4670 |
|
NK-cells |
D21 |
Pt.#2_D21_PBMC_IRB20291_MDIPA_10-13-2021_1 |
7353 |
|
NK-cells |
D28 |
Pt.#2_D28_PBMC_IRB20291_MDIPA_10-13-2021_1 |
5770 |
|
Early NK |
Screening |
Pt#2_Screening_PBMC_IRB20291_MDPA_11-10-2021_1_concat |
3623 |
|
Early NK |
D4 |
Pt.#2_D4_PBMC_IRB20291_MDIPA_10-13-2021_1 |
3130 |
|
Early NK |
D8 |
Pt2_D8_PBMC_merge1 |
4462 |
|
Early NK |
D14 |
Pt.#2_D14_PBMC_IRB20291_MDIPA_10-13-2021_1 |
1841 |
Although changes in the percentage of total CD4+ T cells differed among the two patients, both patients showed an increase in CD4+ TEMRA cells, while instead an increase in CM and EM CD4+ T cells was only present in the second patient (Fig. 2D and Sup. Fig. 2H-J).
An increase in pro-inflammatory CD16+ cells, such as intermediate and non-classical monocytes, was additionally observed in both patients, but the classical monocyte population remained almost unchanged (Sup. Fig. 3A-G). In line with our observation that leflunomide appears to generally increase anti-viral T cell responses, we also noted a consistent increase among the two patients in total NKT (Sup. Fig. 3H-I) and TCR yδ cells (Sup. Fig. 3J-K), which play a pivotal role in countering viral infection14. A robust increase in the NK cell population, specifically in mature NK cells, over the course of the study was noted in the first patient (Sup. Fig. 4A-E).
Consistent with the decrease in the B cell population, the immunoglobulin profile showed a decrease in total IgM levels in the first patient (Sup. Fig. 5A), an effect that was previously associated with a relatively rapid recovery from COVID-1915. This effect was observed seven days after the patient began leflunomide treatment, while total IgG levels remained mostly unchanged (Sup. Fig. 5B). A decrease in IgM levels was instead not observed in the second patient (Sup. Fig. 5C-D).
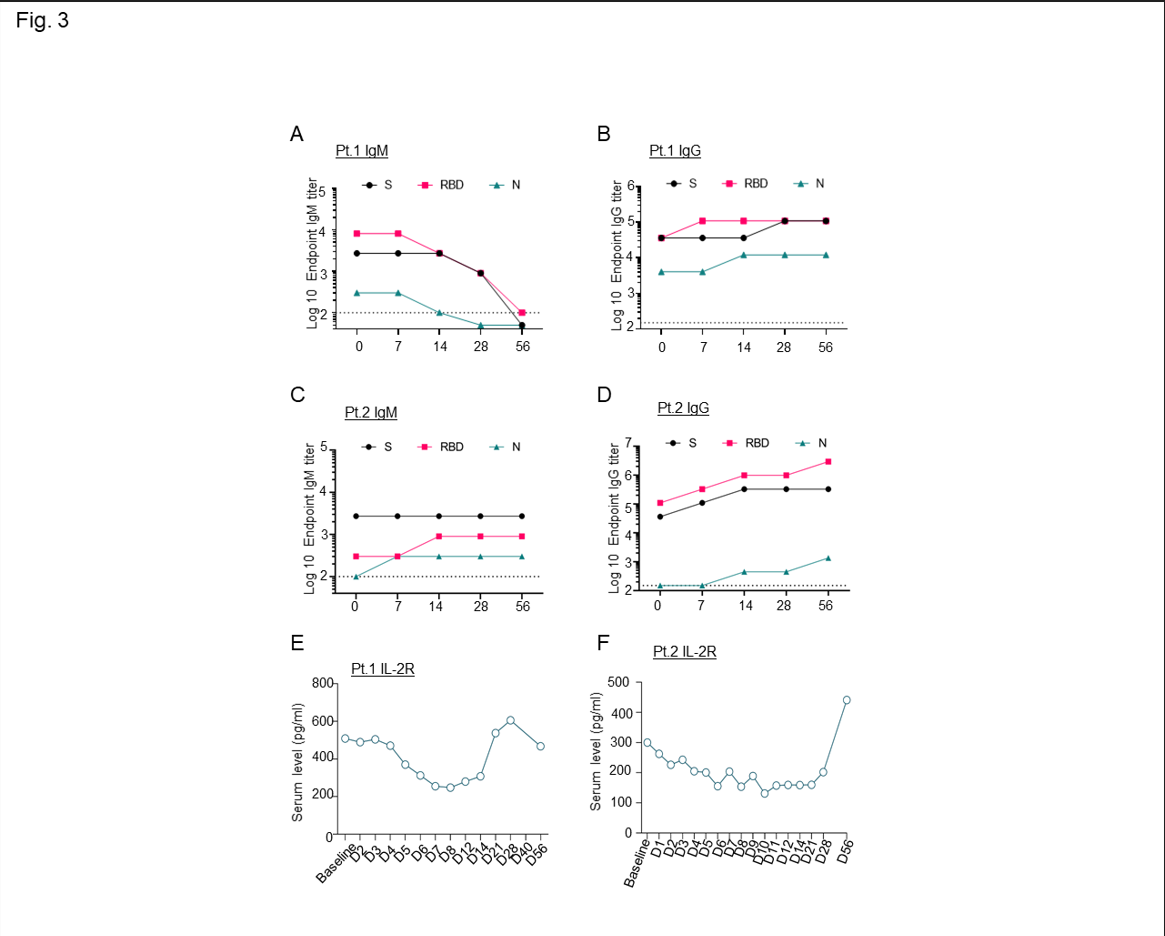
Supplementary Figure 5: A-B-C-D) Longitudinal evaluation of serum IgM (A,C) and IgG (B,D) immunoglobulin reactive to SARS-CoV-2 nucleoprotein (N), receptor-binding domain (RBD), and spike (S) collected from the patients at baseline (time zero), through the course of leflunomide treatment (up to 14 days) and follow up (up to 56 days); E-F) Longitudinal evaluation of serum cytokines IL-2R through the treatment course (14 days) and follow up appointments (up to day 56).
A cytokine array showed that leflunomide treatment decreased soluble IL2R (Sup. Fig. 5E-F); high levels are regularly associated with inhibition of CD8+ cytotoxic T cells and high rates of hospitalization and mortality16.
Consistent with previously published data, here we show that leflunomide affects B cell proliferation17, and for the first time report that leflunomide stimulates anti-viral activity by promoting innate immunity. Our patients treated on a leflunomide protocol had rapid improvements that were coupled with temporal and favorable changes in immunologic response. An important limitation is the small number of patients enrolled to date; however, the data align with two pilot studies comparing leflunomide to leflunomide plus institutional standard of care, one showing that leflunomide plus standard of care conferred favorable SARS-CoV-2 clearance and hospital discharge10, and the other demonstrating a shorter duration of viral shedding and a reduction in C-reactive protein18. A third study assigned patients to either leflunomide or leflunomide plus IFNα and found no statistically significant difference in terms of duration of viral shedding or length of hospital stay19. However, the dosing used was lower than in our study, and it is possible that the enhancement of IFNα signaling by leflunomide precludes additional benefit of IFNα intervention. Finally, two separate case series described patients taking teriflunomide for multiple sclerosis who contracted COVID-19 but experienced self-limiting infection20,21.
Conclusion
We used immunologic correlative experiments to describe the immune response of two patients with severe COVID-19 who were treated with the immune modulator leflunomide. Further study is warranted to more definitely address the effectiveness of leflunomide in the treatment of COVID-19, especially in patients with cancer. Leflunomide treatment might also be relevant for patients who are immunocompromised due to factors other than cancer, but additional evidence is needed. With ongoing COVID-19 transmission, which can overcome precautions such as hand washing and social distancing,22 and occurrence of breakthrough infections in vaccinated individuals, therapeutic agents that target both the virus and host inflammatory response would be helpful despite the availability of currently approved anti-viral agents. Furthermore, from an access to care perspective, especially in resource-limited areas, an inexpensive, readily available, effective drug with existing safety data in humans is relevant in the real-world setting.
Acknowledgments
We are grateful to the Pathogen & Microbiome Division at TGen North for the sequencing of SARS-CoV-2 strains. The Biostatistics Core at City of Hope was supported by the National Institutes of Health under award number P30CA033572. This research was also in part supported by a P30CA033572 Supplement. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also acknowledge support from the Norman and Sadie Lee Foundation.
Conflict of Interest
The authors declare that they have no competing interests that are relevant to this study.
References
- Priyanka, Choudhary OP, Singh I. Protective immunity against COVID-19: Unravelling the evidences for humoral vs. cellular components. Travel Med Infect Dis. 2021; 39: 101911.
- Davis JP, Cain GA, Pitts WJ, et al. The immunosuppressive metabolite of leflunomide is a potent inhibitor of human dihydroorotate dehydrogenase. Biochemistry. 1996; 35(4): 1270-1273.
- Baumann P, Mandl-Weber S, Völkl A, et al. Dihydroorotate dehydrogenase inhibitor A771726 (leflunomide) induces apoptosis and diminishes proliferation of multiple myeloma cells. Mol Cancer Ther. 2009; 8(2): 366-375.
- Romano M, Ruggiero A, Squeglia F, et al. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. 2020; 9(5).
- Parekh JM, Vaghela RN, Sutariya DK, et al. Chromatographic separation and sensitive determination of teriflunomide, an active metabolite of leflunomide in human plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2010; 878(24): 2217-2225.
- Xiong R, Zhang L, Li S, et al. Novel and potent inhibitors targeting DHODH, a rate-limiting enzyme in de novo pyrimidine biosynthesis, are broad-spectrum antiviral against RNA viruses including newly emerged coronavirus SARS-CoV-2. bioRxiv. 2020:2020.2003.2011.983056.
- Wang E, Jan AS, Doan VP, et al. Leflunomide therapy for refractory cytomegalovirus infections in hematopoietic stem cell transplant recipients. J Oncol Pharm Pract. 2019; 25(7): 1731-1737.
- Williams JW, Javaid B, Kadambi PV, et al. Leflunomide for polyomavirus type BK nephropathy. N Engl J Med. 2005; 352(11): 1157-1158.
- Rosenzweig M, Palmer J, Tsai NC, et al. Repurposing leflunomide for relapsed/refractory multiple myeloma: a phase 1 study. Leuk Lymphoma. 2020; 61(7): 1669-1677.
- Wang Q, Guo H, Li Y, et al. Efficacy and Safety of Leflunomide for Refractory COVID-19: A Pilot Study. Front Pharmacol. 2021; 12: 581833.
- Siemasko KF, Chong AS, Williams JW, et al. Regulation of B cell function by the immunosuppressive agent leflunomide. Transplantation. 1996; 61(4): 635-642.
- Breedveld FC, Dayer JM. Leflunomide: mode of action in the treatment of rheumatoid arthritis. Ann Rheum Dis. 2000; 59(11): 841-849.
- Bange EM, Han NA, Wileyto P, et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med. 2021; 27(7): 1280-1289.
- Sabbaghi A, Miri SM, Keshavarz M, et al. Role of γδ T cells in controlling viral infections with a focus on influenza virus: implications for designing novel therapeutic approaches. Virol J. 2020; 17(1): 174.
- Legros V, Denolly S, Vogrig M, et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol. 2021; 18(2): 318-327.
- Jang HJ, Leem AY, Chung KS, et al. Soluble IL-2R Levels Predict in-Hospital Mortality in COVID-19 Patients with Respiratory Failure. J Clin Med. 2021; 10(18).
- van der Heijden EH, Hartgring SA, Kruize AA, et al. Additive immunosuppressive effect of leflunomide and hydroxychloroquine supports rationale for combination therapy for Sjögren's syndrome. Expert Rev Clin Immunol. 2019; 15(7): 801-808.
- Hu K, Wang M, Zhao Y, et al. A Small-Scale Medication of Leflunomide as a Treatment of COVID-19 in an Open-Label Blank-Controlled Clinical Trial. Virologica Sinica. 2020; 35(6): 725-733.
- Wang M, Zhao Y, Hu W, et al. Treatment of Coronavirus Disease 2019 Patients With Prolonged Postsymptomatic Viral Shedding With Leflunomide: A Single-center Randomized Controlled Clinical Trial. Clinical Infectious Diseases. 2020; 73(11): e4012-e4019.
- Maghzi AH, Houtchens MK, Preziosa P, et al. COVID-19 in teriflunomide-treated patients with multiple sclerosis. Journal of Neurology. 2020; 267(10): 2790-2796.
- Mantero V, Baroncini D, Balgera R, et al. Mild COVIDâ19 infection in a group of teriflunomideâtreated patients with multiple sclerosis. Journal of Neurology. 2021; 268(6): 2029-2030.
- Priyanka, Choudhary OP, Singh I, et al. Aerosol transmission of SARS-CoV-2: The unresolved paradox. Travel Med Infect Dis. 2020; 37: 101869.
