Bispecific Antibodies as an Alternative to Antibody Cocktails for SARS-CoV-2: A Mini- Review
Gavin Yuen*, Saundarai Bhanot#, Jeremy Steen, Minahil Syed, Austin Mardon#
Sharpen the Quill, Ontario, Canada
Abstract
Vaccination is a powerful inducer of immunity against SARS-CoV-2 and its recent variants. However, it is important to expand the defensive repertoire against this virus as vaccination is not always efficacious or accessible to everyone. Protein therapeutics in the form of monoclonal antibodies have been used to neutralize the Spike protein, but their efficacy has been limited with rapidly evolving mutations. Cocktail antibodies have been used to combat antigenic escape through diversifying antigen recognition and the overall neutralization capacity. However, the production of cocktail antibodies can be costly and requires a high dosage to achieve the desired therapeutic effect. Alternatively, bispecific antibodies have been used, which contain two recognition specificities within the same molecule. This effectively reduces the cost of production and dosage required to achieve a target therapeutic effect. Bispecific antibodies were reported to bind SARS-CoV-2 antigen with nanomolar affinities. The neutralization potentials (IC50 values) within the same studies were generally more efficacious than their cocktail antibody counterparts. Some studies showed that bispecific antibodies could also confer additional neutralization effector functions, such as recruiting the complement system. Although the recognition of variants was diverse, to our knowledge, there is no data to suggest that bispecific antibodies have a broader recognition of variant strains than cocktail antibodies. Future studies should aim to explore the clinical benefits of bispecific antibodies for SARS-CoV-2 and the emerging variant strains to better understand its benefits in treatment.
Introduction
The emergence of SARS-CoV-2 and its variants have caused severe health, social, and economic burdens.1 Vaccination is one of the most effective defenses, however it may not be enough to end the pandemic due to issues such as inadequate worldwide accessibility, vaccine hesitancy, and the declining effectiveness against variants.2 Effective therapies are essential for unvaccinated populations, immunocompromised individuals who are unable to build protective immunity following immunization, and SARS-CoV-2 patients.3
Throughout the pandemic, neutralizing antibodies have been thoroughly investigated and have proven to be effective against SARS-CoV-2.4 The receptor binding domain (RBD) of SARS- CoV-2 Spike protein has been a prominent target for the creation of neutralizing antibodies due to its critical role in viral infection and accessibility to B cells.4 The Spike protein is essential for infection as it mediates viral attachment to human angiotensin-converting enzyme 2 receptor and subsequent entry into host cells.4 The RBD has less hindrance because it has only two glycosylation sites compared to eight on the N-terminal domain.4 Although several monoclonal antibody (mAb) mixes have been identified that neutralize SARS-CoV-2 by targeting the RBD, the emergence of novel variants have resulted in antigenic escape from antibody recognition.5 SARS-CoV-2 variants such as B.1.1.7 and B.1.351 have evolved Spike protein mutations which can escape neutralization by single antibodies.6 As a result, cocktail antibodies have gained interest for expanding the neutralizing coverage of such variants.6 The use of bispecific antibodies (bsAbs) is another emerging technique which provides potential in its applications as a SARS-CoV-2 therapeutic.7
Studies using SARS-CoV-2-infected mouse and hamster models have demonstrated the efficacy of antibody cocktails for prophylactic and post-infection treatments.8 Structural analysis by cryo-electron microscopy has further revealed sets of non-overlapping epitopes for SARS-CoV-2 and suggests potential for their combination in antibody cocktail-based therapeutics.8 Despite such potential in antibody cocktails, preliminary research has suggested that one bsAb may be up to 100-fold more potent than parent monoclonal antibodies.7
The administration of antibodies directed against a specific antigen to compensate for the lack of recognition is known as passive immunotherapy (eg. neutralizing mAbs.) Conversely, therapies that aim to elicit effector functions are classified as active immunotherapy (eg. vaccines.)9 BsAbs combine the specificities of two distinct antibodies, allowing the recognition of two distinct epitopes within a single molecule (Figure 1a.)7 With recent scientific advances, bispecific technologies have become established therapeutics in the context of cancer immunotherapy.9 This is important to investigate in the context of SARS-CoV-2 because there may not necessarily be an overlap of antigen specificity from patient to patient, so the administration of a low dosage of bsAbs rather than a large dosage of cocktail antibodies is ideal to scavenge a broad range of antigen.7
While existing literature has explored the use of mAbs and antibody cocktails for SARS-CoV-2 related treatments, the efficacy of these monoclonal antibodies and antibody cocktails in comparison to alternative bsAbs remains limited.10 The proposed method of utilizing bsAbs provides immense promise as an alternative technique in the context of combating SARS-CoV-2.11
Binding Affinities to SARS-CoV-2 Antigen
The recognition of pathogens by bsAbs make them an attractive potential alternative to mAb cocktails. While naturally occurring antibodies typically target one antigen, the development of bsAbs has allowed for the simultaneous recognition of two epitopes (Figure 1b.)12 In particular, the binding affinity of a ligand for its target can be evaluated using the equilibrium dissociation constant value (KD), where a smaller KD value indicates a greater binding affinity to the target and reflects high avidity. Several preclinical studies show effective binding strengths of bispecific antibodies (referenced in Table 1).
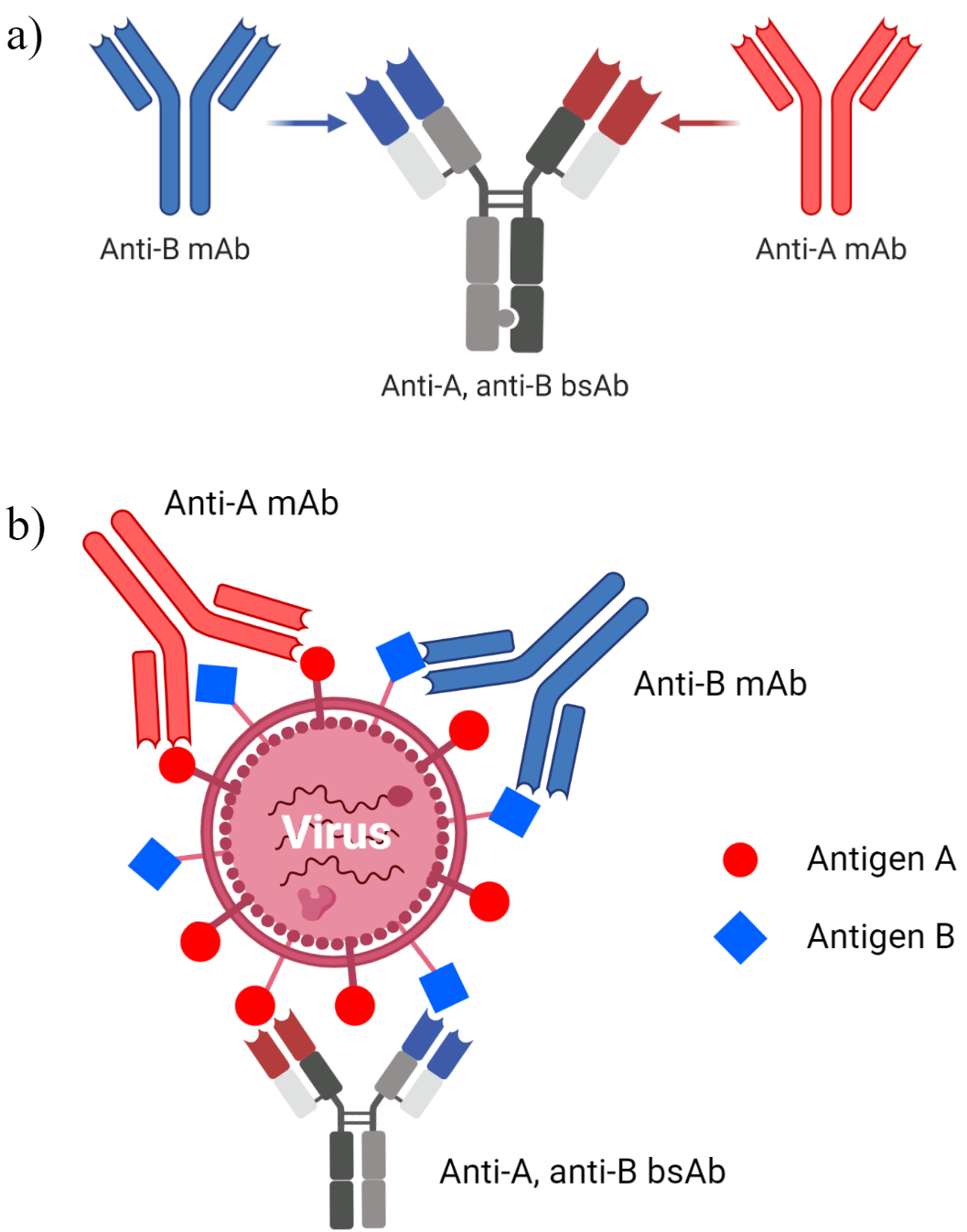
Figure 1: General Illustration of Monoclonal Antibody Cocktail Compared to Bispecific Antibodies. a) Bispecific antibody design for hypothetical antigens A and B. b) Comparison of binding recognition and stoichiometries between antibody cocktails and bsAbs. Figure created using BioRender
Table 1: Reported Dissociation Constant (KD) Values of Developed BsAbs
|
Author, Year Published |
Type of Study |
Type of SARS-CoV-2 Virus Used |
bsAb KD (nM) |
|
Gasparo et al., 202113 |
In vivo and In vitro |
Pseudotype |
CoV-X2 and Spike protein - 0.18 CoV-X2 and RBD - 2.35 |
|
Lim et al., 202114 |
In vitro |
Pseudotype and Authentic |
RBD KDapp* Bis1 (VH A01/Fab C01) <0.001 Bis2 (VH B01/Fab C01) <0.001 Bis3 (VH A01/Fab D01) <0.001 Bis4 (VH B01/Fab D01) <0.001
Spike full ectodomain KDapp* Bis1 (VH A01/Fab C01) 0.603 Bis2 (VH B01/Fab C01) 0.611 Bis3 (VH A01/Fab D01) 0.395 Bis4 (VH B01/Fab D01) 0.127
*All above values are apparent (KDapp) - as derived from a 1:1 binding model of the data |
|
Dong et al., 202015 |
In vitro |
n/a |
1B-3F (Humanized llama antibody)- 0.25 |
|
Dong et al., 202016 |
In vitro |
Pseudotype |
1B-3F (Humanized llama antibody)- 0.25 (comparison to following tri-specific antibodies: - 3F-1B-2A (KD ~ 0.047) - 1B-3F-2A (KD ~ 0.095)
|
|
Wu et al., 202117 |
In vivo and In vitro |
Live, Pseudotype, and Variants
|
Nb15-NbH-Nb15 and RBD - 0.54 Nb15-NbH-Nb15 and HSA - 7.7 |
A bsAb termed CoV-X2 was developed from two antibodies obtained from individuals post recovery from SARS-CoV-2.13 Avidity data showed that the binding of one arm of the bsAb could improve the intermolecular interactions of the other arm due to proximity. Thus, while mAbs have avidity only at high concentrations of receptor binding domains, CoV-X2 was found to have a high avidity reflected through dissociation rates at all concentrations13
With respect to specificity for certain domains of SARS-CoV-2, antibodies targeting non- overlapping sites were found to be the most potent.7 As a result, the RBD and N-terminal domain have been of particular interest in antibody recognition. Currently, the development of VH/Fab antibodies targeting Spike proteins have shown that both sites are able to inhibit the binding of the ACE-2 receptor to the S protein.14 Another advantage of two epitope recognition by bsAbs, and potentially tri-specific antibodies (Table 1), is the ability to neutralize more pathogens with a lower dose compared to cocktail antibodies. 14 Thus, the production of lower bsAb quantities is more economic relative to cocktail antibodies. In terms of clinical significance, there is currently no data to compare the toxicities of bsAb dosages relative to cocktail antibodies in patients.
BsAb Neutralization Potency Compared to Cocktail Antibody Counterparts
The neutralization potential of bsAbs against SARS-CoV-2 is reflected by their IC50 values. As IC50 is dependent on the concentration of the substrate, only comparisons within the same study were made (Table 2).7,13,14,17-20 In general, it was observed that the bsAbs had lower IC50 values than their respective monovalent antibodies or cocktail combination of these monovalent antibodies, which indicated a more robust neutralization potential.7,14,18,19 Two studies in particular looked into bsAbs similar to IgG, which neutralized the virus with lower IC50 values compared to their respective mAbs and cocktail antibodies.13,14 A potential explanation was the improved affinity, binding mechanism, and scaffold which allowed bsAbs to adopt multiple conformations to bind epitopes of concern.14 In other words, these epitopes were more easily accessed when bsAbs were used because of the improved orientation of the binding region towards the antigen.14 In addition, since these studies used IgG antibodies, Fc modifications could be made to boost effector functions.13 The CoV-X2 was found to have LALA-PG mutations in the Fc region, which enabled Fc interactions with its receptors to activate the complement system in response to infection while avoiding adverse effects.13 The diversity of structure and potential for modification in bsAbs allows for more effective neutralization than standard antibody therapies.
Table 2: IC50 Values for Select Antibodies Against SARS-CoV-2 Epitopes.
|
Pseudotype SARS-CoV-2 Neutralization |
Authentic SARS-CoV-2 Neutralization |
|||
|
Authors |
Monovalent Antibody IC50 (μg/mL) |
bsAb IC50 (μg/mL) |
Monovalent Antibody (μg/mL) |
bsAb IC50 (μg/mL) |
|
Li T., et al.18 |
Sybodies (synthetic nanobodies)
MR3: 0.42
MR17: 13.18
LR5: 2.10 |
Bispecific nanobodies
MR3-MR3[13 GS]: 0.133
MR3-MR3[19 GS]: 0.121
MR3-MR3[24 GS]: 0.114
MR3-MR3[34 GS]: 0.010
MR17m-MR17m[13 GS]: 0.029
MR17m-MR17m[16 GS]: 0.121
MR17m-MR17m[19 GS]: 0.072
MR17m-MR17m[24 GS]: 0.193
LR5-MR3 [13 GS]: 0.112
LR5-MR3 [19 GS]: 0.111
LR5-MR3 [34 GS]: 0.118 |
n/a |
n/a |
|
Bracken C.J., et al.19 |
VHA01: (0.763±0.046)
VHB01: (3.032±0.978)
VHB02: (2.065±0.378)
Cocktails VHA01+B01: (0.400±0.215)
VHA01+B02: (0.491±0.482)
VHB01+B02: (5.608±2.554) |
VH2A01-B01: (0.0214±0.0081)
VH2A01-B02: (0.0306±0.0189)
VH-Fc-A01: (0.209±0.066)
VH-Fc-B01: (0.145±0.019)
VH-Fc-B02: (0.462±0.152)
VH3B01: (0.00690±0.00212) |
n/a |
VH2A01-B01: (0.347±0.087)
VH2A01-B02: (0.743±0.184)
VH-Fc: (2.613±0.647)
VH3B01: (0.176±0.069) |
|
Miao X., et al.20 |
n/a |
89C8-ACE2: 0.029 x 10-5 μM |
n/a |
89C8-ACE2: 1.7 x 10-3 μM |
|
Cho H., et al.7 |
CV503: 0.0029
CV664: 0.0135
CV993: 0.0370 |
n/a |
CV503_521_GS: 1.6 x 10-3
CV521_1182_GS: 4.0 x 10-3
CV503_993_EL: 0.0268
CV1206_521_GS: 1.2 x 10-3
CV521_503_GS: 8.0 x 10-3
CV664_993_GS: 5.1 x 10-3
CV993_521_GS: 0.0261
CV503_664_GS: 3.2 x 10-3
CV503_664_EL: 3.0 x 10-3
CV1206_521_GS: 0.0160 |
CV503_521_GS: 0.20 x 10-3
CV521_1182_GS: 0.50 x 10-3
CV503_993_EL: 1.90 x 10-3
CV1206_521_GS: 0.5 x 10-4
CV521_503_GS: 0.45 x 10-3
CV664_993_GS: 3.33 x 10-3
CV993_521_GS: 1.36 x 10-3
CV503_664_GS: 4.17 x 10-3
CV503_664_EL: 0.21 x 10-3
CV1206_521_GS: 1.01 x 10-3 |
|
Wu X., et al.17 |
1 × Nb15: 0.3074 ± 0.0237
2 × Nb15: 0.0003 ± 0
3 × Nb15: 0.0004 ± 0
4 × Nb15: 0.0002 ± 0.0001 |
Nb15-NbH: 0.5529 ± 0.0889
NbH-Nb15: 0.1974 ± 0.004
Nb15-Nb15-NbH: 0.0251 ± 0.0058
NbH-Nb15-Nb15:0.0008 ± 0.0001
Nb15-NbH-Nb15: 0.0004 ± 0
Nb15-Fc: 0.0009 ± 0.0001 |
n/a |
n/a |
|
Gasparo R.D., et al.13 |
C121, C135: higher IC50 than CoV-X2 (estimated from data) |
CoV-X2: 4 x 10-5 μM |
n/a |
CoV-X2: 9 x 10-4 μM |
|
Lim S.A., et al.14 |
*brackets are 95% CI
VH-Fc A01: 0.23 (0.13–0.40)
VH-Fc B01: 0.16 (0.10–0.27)
IgG C01: â«15
IgG D01: â«15
Cocktails VH-Fc A01 + IgG C01: 0.75 (0.52–1.09)
VH-Fc B01 + IgG C01: 0.58 (0.39–0.86)
VH-Fc A01 + IgG D01: 0.26 (0.19–0.35)
VH-Fc B01 + IgG D01: 0.37 (0.27–0.50) |
Bis1 (VH A01/Fab C01): 0.78 (0.39–1.94)
Bis2 (VH B01/Fab C01): 0.92 (0.53–1.82)
Bis3 (VH A01/Fab D01): 0.015 (0.009–0.023)
Bis4 (VH B01/Fab D01): 0.012 (0.009–0.016) |
VH-Fc A01: 2.04 (1.50–2.88)
VH-Fc B01: 2.38 (2.12–2.68) |
Bis1 (VH A01/Fab C01): 1.16 (0.95–1.40)
Bis2 (VH B01/Fab C01): 1.10 (0.77–1.61)
Bis3 (VH A01/Fab D01): 0.11 (0.08–0.17)
Bis4 (VH B01/Fab D01): 0.14 (0.09–0.20) |
In three other studies, nanobodies were used which similarly yielded relatively low IC50 values in the bispecific format compared to their monovalent forms.17-19 It was reported that Fc domains often led to unwanted antibody-dependent enhancement of infections.17 Therefore, these nanobodies were constructed without the Fc domains and were observed to have effective viral neutralization because of their ease of penetration at the site of infection.17,18 In addition, one of the studies reported that the biparatopic VH2 nanobody constructs demonstrated inter- and intra- RBD avidity that occluded the Spike protein-ACE2 binding interaction, and ultimately outperformed their monovalent and cocktail counterparts.19
Resistance to Mutational Escape
With the growing concern of SARS-CoV-2 variants, it is imperative that antibody recognition is diverse and covers a wide range of epitopes. Several studies that focused on cocktail antibodies demonstrated a diverse range of epitope recognition with just the mAbs. In one study, the reported mAbs iB6, iB14, and iB20 recognized distinct epitopes on the kappa, delta, and lambda variants, which were variants of concern.21 While no experiments were tested using the cocktails, it was hypothesized that since the mAbs recognized different epitopes, there would be increased recognition potential and therefore a greater barrier to mutational escape.21
Another study reported antibody cocktails using REGN mAbs and it was shown to neutralize Spike protein mutations.22 This was reasoned with the ability of different antibodies to cover non- overlapping Spike epitopes.22 However, no variants of concern were tested.22
There is evidence to suggest that variants of concern can be neutralized by bsAbs (Table 3).7,13,23 As with mAb cocktails, the recognition of a diverse array of epitopes and conserved sequences across variants allows for neutralization.7,13,23 Although bsAbs neutralize a broader array of SARS-CoV-2 mutants than individual mAbs, there was a lack of data suggesting that bsAbs performed better than cocktail antibodies in terms of mutational escape.
Table 3: Bispecific Antibody Neutralization Against SARS-CoV-2 Variants
|
Authors |
bsAb or cocktail Ab combinations |
Variants of Concern Neutralization (Yes (Y)/No (N)/Not Tested (NT)) |
|||
|
B.1.617.2 |
B.1.1.7 |
B.1.1351 |
P.1 |
||
|
Gasparo R.D., et al.13 |
CoV-X2 |
NT |
Y |
Y |
Y |
|
Cho H., et al.7 |
CV521_1182_GS |
N |
N |
N |
N |
|
|
CV503_993_EL |
Y |
Y |
Y |
Y |
|
|
CV1206_521_GS |
N |
N |
Y |
N |
|
|
CV521_503_GS |
Y |
Y |
Y |
Y |
|
|
CV664_993_GS |
Y |
Y |
Y |
Y |
|
|
CV993_521_GS |
Y |
Y |
Y |
Y |
|
|
CV503_664_GS |
Y |
Y |
Y |
Y |
|
|
CV503_664_EL |
Y |
Y |
Y |
Y |
|
|
CV1206_521_GS |
Y |
Y |
Y |
Y |
|
White I., et al.23 |
CV19B307 |
NT |
NT |
NT |
Y |
Discussion
The purpose of the present review was to identify the quantity and type of studies investigating the use of bsAbs in the treatment of SARS-CoV-2 as an alternative to cocktail antibodies. While cocktail antibodies allowed for the recognition of multiple epitopes, a more efficient way to target pathogens may be achieved through bsAbs because they reduced the number of molecules required to target different structures. While the literature for pre-clinical trials of bsAbs remains limited, in vitro analysis showed promise in the binding and neutralization of bsAbs to SARS-CoV-2 related epitopes.7, 13-17, 18-20 BsAbs were shown to have nanomolar binding affinities and their IC50 values consistently outperformed their cocktail antibody counterparts, which highlighted an advantage of this therapeutic.7, 13-17, 18-20
Cocktail antibodies have been found to be effective against SARS-CoV-2 when identified using mouse models and in clinical trials using recombinant SARS-CoV-2 RBDs.4 Such naturally occurring antibodies were effective against a single antigen, however their efficiency and potency could be improved through the addition of recognition sequences on the antibodies.4,9 SARS-CoV-2 antigen was shown to be recognized by bsAbs with nanomolar binding affinities, which were extremely strong interactions. This is promising because recognizing SARS-CoV-2-related antigen in the midst of Spike protein mutations allows for the development of novel therapeutics.7 From the bsAb studies identified for the present review, bsAbs generally had lower IC50 values compared to their monoclonal or cocktail antibody counterparts.13,14 This was attributed to structural and functional differences where bsAbs could access multiple epitopes simultaneously with more flexibility.13,14 Additionally, due to their small size and inter- and intra-avidity binding, bispecific nanobodies were found to have improved tissue penetration when compared to monovalent nanobodies.17-19 With greater access to epitopes, the overall viral neutralization was increased.17-19 Future studies that modify Fc domains could incorporate the complement system for greater viral clearance.13 Alternatively, efforts that modify bsAb recognition towards natural killer or T cells could mediate cross-protection and confer additional protection upon re-exposure.
Studies showed that both cocktail antibodies and bsAbs could neutralize a variety of Spike protein mutations for SARS-CoV-2. 7,13,21-23 Specifically, these antibodies could recognize conserved regions between mutants, which resulted in neutralization. In addition, data suggested that cocktail antibodies had the potential to suppress variants of concern.7,13,23 While studies suggested that bsAbs were able to neutralize B.1.617.2, B.1.1.7, B.1.1351, and P.1 variants of concern, only some bsAbs were able to suppress all four variants.7,13,23 Overall, there was insufficient evidence to suggest bsAbs would outperform cocktail antibodies in terms of recognizing and neutralizing variants. This may pose as a potential limitation if new bsAbs need to be developed every time Spike mutations evade their full recognition capacity. Future studies could aim to further broaden the recognition specificities of bsAbs by targeting semi-conserved antigens across the mutants to improve the recognition diversity. This would challenge mutational escape in clinical settings and potentially reduce the number of viral infections. Polyclonal passive immunotherapy could also be employed to provide for immediate defense and diversified antigen recognition. Another limitation of bsAbs is the potential lack of persistency in the upper and lower respiratory tracts due to low half-lives.7 Future works in animal models are required to understand and optimize the pharmacokinetics of bsAbs, relative to existing mAb therapies.
The development of bsAbs for SARS-CoV-2 holds potential therapeutic benefit. Pre-clinical data indicated that bsAbs had strong binding affinities to SARS-CoV-2 antigen and that there was effective neutralization of the wild-type and mutant strains. Future in-vivo and in clinical studies should continue to examine these properties and whether the virus and its mutant strains relapse after bsAb treatment. Further modifications to improve the range of bsAb neutralization mechanisms should be welcomed to diversify the killing repertoire against emerging SARS-CoV-2 strains.
Conclusion
The present review involved a broad search in the literature to identify the quantity and type of studies investigating the use of bsAbs in the treatment of SARS-CoV-2 as an alternative to cocktail antibodies. We identified three major themes constituting the binding, neutralization potential and resistance to mutational escape of bsAbs. Many reported bsAbs had nanomolar binding dissociation constants and micro- and nanomolar IC50 values, which were more efficacious than their respective mAb cocktail counterparts. This presents a more economic and powerful method of recognizing SARS-CoV-2 antigen. The development of bsAbs and their subsequent applications in clinical studies can prove to be an efficient treatment for individuals who are unable to develop sufficient natural antibodies through vaccines.
Acknowledgment
The authors would like to thank Sharpen the Quill for their guidance throughout this project and acknowledge TakingITGlobal for their $750 RisingYouth Grant to make this project possible.
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- Osterrieder A, Cuman G, Pan-Ngum W, et al. Economic and social impacts of COVID-19 and public health measures: results from an anonymous online survey in Thailand, Malaysia, the UK, Italy and Slovenia. BMJ Open. 2021; 11: e046863. doi: 10.1136/bmjopen-2020-046863.
- Ian White, Ninkka Tamot, Rajitha Doddareddy, et al. Bifunctional molecules targeting SARS-CoV-2 spike and the polymeric Ig receptor display neutralization activity and mucosal enrichment, mAbs. 2021; 13: 1, doi: 10.1080/19420862.2021.1987180
- Ljungman, P. Vaccination of immunocompromised patients. Clinical Microbiology and Infection. 2012; 18: 93-99. doi:10.1111/j.1469-0691.2012. 03971. X.
- Dong J, Zost SJ, Greaney AJ, et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat Microbiol. 2021; 6, 1233–1244. doi: 10.1038/s41564-021-00972-2.
- Alejandra, Tortorici M, Beltramello Martina, et al. Ultrapotent Human Antibodies Protect against SARS-CoV-2 Challenge via Multiple Mechanisms. Science. 2020; 370 (6519): 950–57. doi:10.1126/science.abe3354.
- Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021; 184(9): 2384-2393.e12. doi: 10.1016/j.cell.2021.03.036.
- Hyeseon C, Kay G-WK, Deli H, et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci Transl Med. 2022;13(616): eabj5413. doi:10.1126/scitranslmed.abj5413.
- Su S-C, Yang T-J, Yu P-Y, et al. Structure-guided antibody cocktail for prevention and treatment of COVID-19. PLoS Pathog. 2021; 17(10): e1009704. doi: 10.1371/journal.ppat.1009704.
- Papaioannou NE, Beniata O V, Vitsos P, et al. Harnessing the immune system to improve cancer therapy. Ann Transl Med. 2016; 4(14). doi: 10.21037/atm.2016.04.01
- Zhang J, Zhang H, Sun L. Therapeutic antibodies for COVID-19: is a new age of IgM, IgA and bispecific antibodies coming? MAbs. 2022;14(1): 2031483. doi: 10.1080/19420862.2022.2031483
- Cho H, Gonzales-Wartz KK, Huang D, et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci Transl Med. 2021;13(616): eabj5413. doi:10.1126/scitranslmed.abj5413/.
- Jia L, Liu Y-P, Tian L-F, et al. Potent neutralizing RBD-specific antibody cocktail against SARS-CoV-2 and its mutant. MedComm. 2021; 2(3): 442-452. doi: 10.1002/mco2.79.
- De Gasparo R, Pedotti M, Simonelli L, et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature. 2021; 593(7859): 424-428. doi:10.1038/s41586-021-03461-y.
- Lim SA, Gramespacher JA, Pance K, et al. Bispecific VH/Fab antibodies targeting neutralizing and non-neutralizing Spike epitopes demonstrate enhanced potency against SARS-CoV-2. MAbs. 2021; 13(1): 1893426. doi:10.1080/19420862.2021.1893426.
- Dong J, Huang B, Jia Z, et al. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg Microbes Infect. 2020; 9(1): 1034-1036. doi:10.1080/22221751.2020.1768806.
- Dong J, Huang B, Wang B, et al. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci Rep. 2020;10(1):17806. doi:10.1038/s41598-020-74761-y.
- Wu X, Cheng L, Fu M, et al. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep. 2021;37(3): 109869. doi: 10.1016/j.celrep.2021.109869.
- Li T, Cai H, Yao H, et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat Commun. 2021;12(1):4635. doi: 10.1038/s41467-021-24905-z.
- Bracken CJ, Lim SA, Solomon P, et al. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat Chem Biol. 2021; 17(1): 113-121. doi:10.1038/s41589-020-00679-1.
- Miao X, Luo Y, Huang X, et al. A novel biparatopic hybrid antibody-ACE2 fusion that blocks SARS-CoV-2 infection: implications for therapy. MAbs. 2020; 12(1): 1804241. doi: 10.1080/19420862.2020.1804241.
- Gorchakov AA, Kulemzin SV, Guselnikov SV, et al. Isolation of a panel of ultra-potent human antibodies neutralizing SARS-CoV-2 and viral variants of concern. Cell Discov. 2021; 7(1): 96. doi: 10.1038/s41421-021-00340-8.
- Baum A, Fulton BO, Wloga E, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020; 369(6506): 1014-1018. doi:10.1126/science.abd0831.
- White I, Tamot N, Doddareddy R, et al. Bifunctional molecules targeting SARS-CoV-2 spike and the polymeric Ig receptor display neutralization activity and mucosal enrichment. MAbs. 2021; 13(1): 1987180. doi: 10.1080/19420862.2021.1987180.
