SARS-CoV-2: Insights from the Immunopathogenesis and Current Clinical Diagnosis and Therapeutic Strategies
Shruti Sharma1*, Ujjawal Sharma2#, Anupama Chaudhary3, Manisha Naithani4, Priyanka Naithani5, Saurabh Prashar2, Bunty Sharma6, Pramod Kumar Nagar7, Prudhvi Lal Bhukya8, Unnati Bhalerao8, Meenakshi Singh9, Manjita Srivastava8, Muneesh Kumar Barman10, Sampan Attri2, Jitender Gairolla2,11#
1Department of Experimental Medicine and Biotechnology, Postgraduate Institute of Medical Education and Research, Chandigarh, India
2Department of Community Medicine and School of Public Health, Postgraduate Institute of Medical Education and Research, Chandigarh, India
3Orinin BioSystems, Karnal, India
4Department of Biochemistry, All India Institute of Medical Sciences, Rishikesh, India
5Department of Pharmacology, Postgraduate Institute of Medical Education and Research, Chandigarh, India
6Chitkara School of Health Sciences, Chitkara University, Punjab, India
7Hematology Oncology Unit, Advance Pediatric Centre, Postgraduate Institute of Medical Education and Research, Chandigarh, Chandigarh, India
8National Institute of Virology, Pune, India
9Transplant Immunology and Immunogenetics Lab (HLA), ACTREC, Tata Memorial Centre
10Laboratory for HIV Research, National Centre for Cell Sciences, Pune-India
11Department of Microbiology, All India Institute of Medical Sciences, Rishikesh, India
Abstract
The global public health scenario is worsening gradually as the confirmed cases of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) infections are incessantly escalating with every passing day. The pathological condition caused by SARS-CoV-2 is termed as Coronavirus disease 2019 (COVID-19). The understanding of SARS-CoV-2 transmission dynamics, immunopathogenesis, and the need for early-stage diagnosis and the effective therapeutic regime are the few immediate challenges faced by healthcare professionals worldwide. More specifically, the role of SARS-CoV-2 in the host’s immunopathogenesis response is crucial to determine the disease severity and its clinical outcome in COVID-19 patients. In the present review, we provide insights into the SARS-CoV-2 pathology, host immune responses including innate, cellular, and humoral responses, and immunomodulatory functions of SARS-CoV-2 including cytokine storm and immune evasion. We also shed light upon the present clinical and laboratory-based applications enrolled in the SARS-CoV-2 diagnosis. Taking into consideration the pathogenesis and SARS-CoV-2 immune function, in the present review, we finally provide succinct insights into the SARS-CoV-2 transmission dynamics, immunopathogenesis, with the assessment of the current diagnostic and preventive/ therapeutic strategies.
Introduction
We are witnessing unprecedented times with the rapidly evolving global COVID-19 pandemic that has brought the world to a socio-economic halt and greatly impacted public health worldwide. The onset of the COVID-19 outbreak was marked in late December 2019, with the reporting of a group of patients suffering from a Severe Acute Respiratory Syndrome-like disease in Wuhan, China. Preliminary investigations revealed that the causative agent is a previously unknown virus and therefore it was named as ‘Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), by the ICTV (International Committee on Taxonomy of Viruses)1. Coronaviruses belong to the Coronoviridae family that comprises of viruses which are characteristically identified by a ribonucleic acid core surrounded by a protein-spiked envelope and their structural resemblance to that of a crown (crown i.e., corona in Latin) has resulted in their nomenclature as coronaviruses2. Coronaviruses (CoVs) form a major group of viruses under the Nidovirales order that comprises three families viz. Arteriviridae, Roniviridae, and Coronaviridae. Most coronaviruses are known to produce mild respiratory and occasionally gastrointestinal-related symptoms with the severity of respiratory disease ranging from mild symptoms such as common cold to even severe respiratory complications such as pneumonia3.
The earliest case of a common cold due to infection by a coronavirus was reported in 19604. Later in 2001, almost with a gap of four decades, a Canadian study reported flu-like symptoms in about 500 patients, wherein 17-18 cases were found to be infected by a coronavirus. Hence, coronavirus infections were inferred as non-lethal until 2002. However, the outbreak of SARS coronavirus (SARS-CoV) in 2003 attributed to a high mortality rate5,6. Furthermore, another serious coronavirus outbreak was occurred in 2012, in Saudi Arabia due to a new coronavirus named Middle East Respiratory Syndrome Coronavirus (MERS-CoV)7. After a gap of seven years, a novel coronavirus struck the world again in December 2019, in a different form and identity viz. the SARS-CoV-2. It was traced to be of zoonotic origin with the seafood and live animal market in Wuhan, China being the prime suspected source. On March 11, 2020, the WHO (World Health Organization) declared the outbreak of Coronavirus Disease-2019 (COVID-19) a ‘pandemic’8, with the causative agent, SARS-CoV-2, defying the continental boundaries and spreading globally and North America and Europe being important and vastly affected epicenters. Evidence-based knowledge on COVID-19 has become crucial to get a better understanding of the etiology, immunopathogenesis, and immunomodulatory activities. Timely assessment of these aspects of the SARS-CoV-2 infection could enable us to build efficient clinical diagnostic and preventive/therapeutic regimes against the ongoing fight against this highly transmissible virus. In this review, we shed light upon the SARS-CoV-2 infection/transmission dynamics, hosts innate and acquired immune responses, and its immunomodulatory functions including the commonly witnessed cytokine storm and immune evasion. Furthermore, we discuss the scope of present clinical diagnostic approaches and preventive/ therapeutic strategies. The evidence-based new knowledge, as succinctly reviewed in the present report, could essentially pave the way for better and effective diagnosis, management, and prevention of ongoing COVID-19 pandemic.
SARS-CoV-2, Outbreak, and Transmission Dynamics
The first few cases of SARS-CoV-2 infected cases identified infections were reported from in Wuhan City China (in the Hubei Province), with symptoms like province of China and these cases presented with an acute respiratory syndrome of idiopathic etiology9. An early outbreak of SARS- Cov-2 infection showed symptoms such as fever, sore throat, and significant dyspnea10,11. On February 11, 2020, WHO named the disease as ‘COVID-19’, and by the end of February, COVID-19 had spread to over 25 nations12,13. An early investigative study performed on the confirmed COVID-19 cases from Wuhan reported the exposure history of the patients to the seafood market11. Besides the live-food, the possibility of seafood contamination with bats or bat droppings was also greatly postulated. Initial phylogenetic analyses of SARS-CoV-2 suggested its resemblance to the beta-coronaviruses, however, the distinctive phylogeny of the RNA-dependent RNA polymerase of SARS-CoV-2 indicated that it was a novel strain of the genus Betacoronavirus and subgenus Sarbecovirus. The genomic analyses revealed an 88% sequence identity of SARS-CoV-2 with bat coronaviruses11,14, and therefore affirmed its zoonotic origin with bats serving as reservoirs for SARS-CoV-2, which further transmit the disease to humans and other animal species. Prior to the WHO’s declaration of the ‘COVID-19 pandemic’ in March 2020, an unexpected rise in the numbers of confirmed COVID-19 positive cases were observed in China. An early report from China revealed that the basic reproduction number (R0) of COVID-19 ranges from 2.2 to 3.58 which was relatively lesser than SARS but higher than MERS15. Of note, the SARS-CoV-2 median incubation duration was found to be 6.4 days (Credible interval (CI)-95%, 5.6 to 7.7 days) and these were very similar to those for MERS and SARS16,17. The key pathology factors such as the onset of symptoms and rate of mortality were found to be dependent on age, pre-conditions (hypertension, diabetes, and cardiovascular disease) and the immune state of the infected individual13,16,18,19. Therefore, a higher susceptibility, severity, and mortality rate of COVID-19 was reported in individuals who were either aged (median > 59 years), or had had aforementioned comorbidities19,20. In the early outbreak, the mortality rate was reported to be between 1.5- 3% in Wuhan, China, however, these statistics largely found to depend on the criteria of diagnosis, the scale of COVID-19 transmission, and geographical location as well10,21. By date, the COVID-19 outbreak has spread to over 208 countries, and the Centre for Disease Control and Prevention (CDC) and WHO are jointly engaged in closely monitoring the situation to forecast its timely impact on public health. As of March 20, 2021, a global death count of 2,694,094 with a total 121,969,223 confirmed cases have been estimated and as a result of the COVID-19 pandemic (https://covid19.who.int/). While assessing the SARS-CoV-2 transmission dynamics, the person-to-person contact established either through direct contact or droplets released in the air (by coughing or sneezing) was found to be the most common route of its transmission to a healthy person13. While, the indirect routes of transmission were also observed, suggesting that SARS-CoV-2 transmission can also occur by touching a surface or fomites (copper, plastic, steel, cardboard, etc.) that contains the existing virus particles22,23. Of note, the half-life of SARS-CoV-2 varies with environmental conditions, and it was notably high on cardboard (~ 24 hours), but found to be at a minimum in aerosols (≤ 4 hours)23.
SARS-CoV-2 Infection, Host Cell Entry, and Immunological Triggers
The 50-200 nanometers (nm, diameter) virion of SARS-CoV-2 is found to be composed of 4 structural proteins viz. spike (S) envelope (E), membrane (M), and nucleocapsid (N) (Fig. 1A)24,25. Out of these, the S protein is essential for attachment of the virus to the membrane of host cells and to gain access into the cells (Fig. 1B)24. Initial validations conferred that SARS-CoV-2 acquires a greater binding to ACE2 protein in humans which was later identified to be its binding receptor by which it ensures entry inside the cell26,27. Of note, processing of S-protein by host cell transmembrane proteases serine 2 (TMPRSS2) was found to be crucial for S-protein- ACE2 coupling, wherein the TMPRSS2 protease plays an activator function and facilitates the SARS-CoV-2 entry into the host cell (Fig. 1B)27. Markedly, this processing of S-protein before its binding to the ACE2 receptor includes its enzymatic modification leading to its cleavage and change in conformation by protease activity of TMPRSS2 that resides in the vicinity of ACE2 receptor i.e., a crucial step in the initiation of viral endocytosis (Fig. 1C)28. Insights from an earlier report analyzing SARS-CoV binding with ACE2 suggest that it enables SARS-CoV-2 internalization and trafficking into the endosomes and then the lysosomes, which subsequently helps in the viral uncoating and fusion (Fig. 1C)29. In this process, the S protein further undergoes a series of post translational modifications including enzymatic cleavage and modifications by both cathepsin L and B, the key lysosomal cysteine proteases, which causes the internalization of the virus into the endo-lysosomes. These steps are crucial in the fusion of viral and late endosome membranes and subsequent discharge of the viral genome (ssRNA) in the host cell cytoplasm (Fig. 1C)30. The latter step is highly pH-dependent, where fusion only occurs after reaching highly acidic conditions in the lysosomes, possessing the utmost cathepsin L activity (Fig. 1C)31. These findings are further supported by the observation that the usage of various lysosomotropic agents inhibits infection by interfering with lysosomal processing of viral membrane proteins, which is a pre-requisite for fusion32. Therefore, it is reasonable to speculate that inhibiting the binding of the S protein to ACE2 or intracellular blocking of its subsequent cleavage by cathepsin L could be the two key target steps in the SARS-CoV-2 infection to develop therapeutics against SARS-CoV-230. Another key therapeutic target could be the Nsp15 protein, which is conserved among coronaviruses and is required for their life cycle and virulence. Nsp15 is a crucial SARS-CoV-2 protein that interferes with the host immune system and facilitates the replication of the virus genome within the host33.
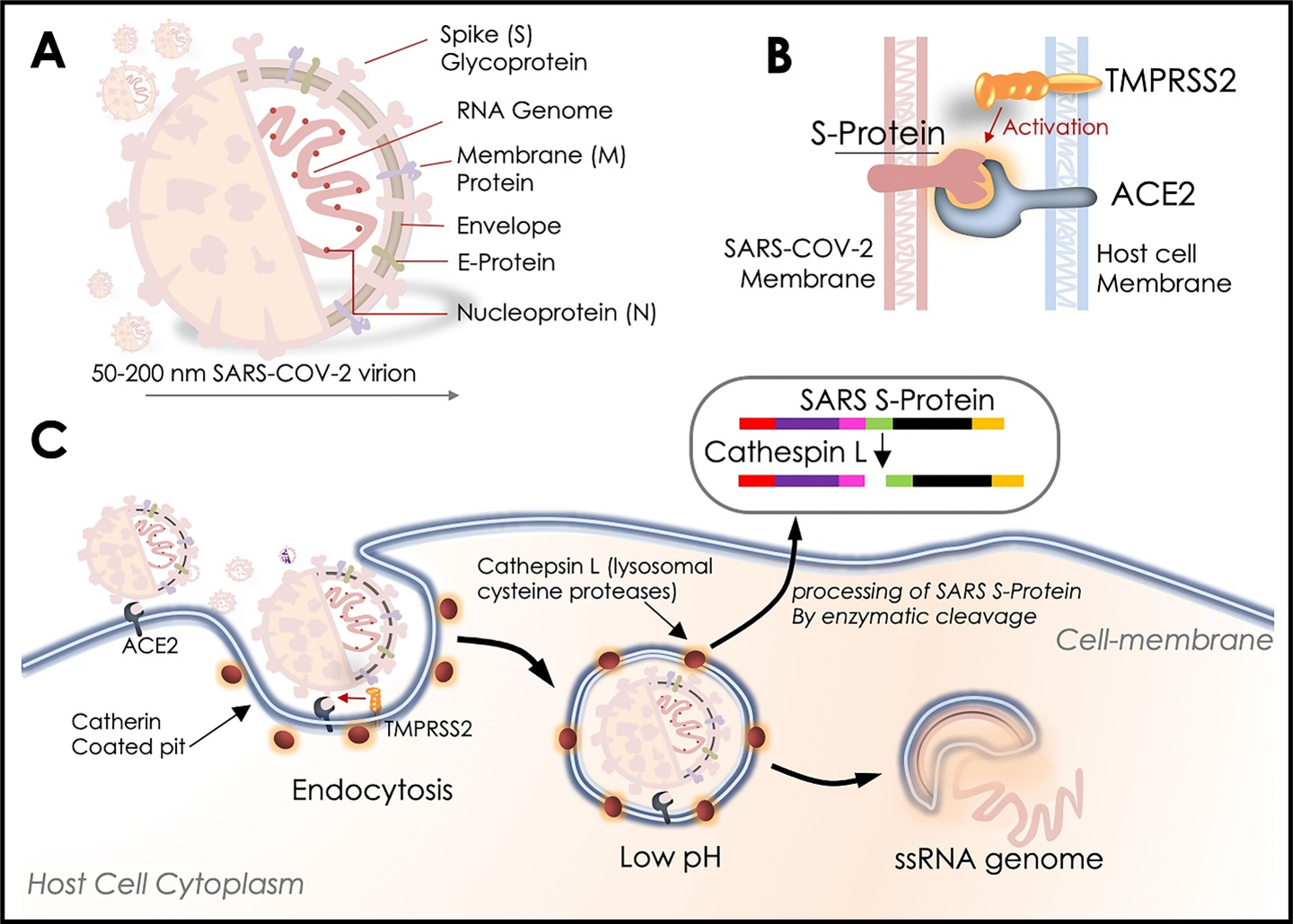
Figure 1. SARS-COV-2, priming of S-protein, and host cell entry. A. SARS-COV-2 virion and its key proteins. B. Schematic diagram showing host cells TMPRSS2-activated binding with SARS-COV-2 S-protein and ACE2 receptor localization at the host cell membrane. C. Schematic diagram mimicking steps for SARS-COV-2 entry into the host cells and enzymatic processing of Spike (S) –protein by host Cathepsin L enzyme before releasing its ssRNA genome to the host cells. This pictorial representation relies on the reference information available for SARS-CoV-2 infection.
Looking at the immunological cues of the SARS-CoV-2’s viral function, frequent lymphopenia, and pneumonia have been observed in infected moderate-to-severe cases34. Of note, SARS-CoV-2 induced alveolar macrophage activity has been suggested to trigger the immunological response in the patient’s lungs that incites cytokines/chemokines release and causes a subsequent activation of T-Cell and the Neutrophils35 (Fig. 2). As a result of this, elevated expressions of pro-inflammatory cytokines viz. TNFα, MCP-1, MIP-1A, IL-2, IL-7, IL-10, and G-CSF were observed in a severely infected cohort of 41 hospitalized COVID-19 patients (Fig. 2)17. These factors suggest a crucial role of cytokine storm leading to viral sepsis and inflammation-induced lung injury. These risks may further promote pneumonitis and acute respiratory distress syndrome (ARDS) that may lead to the organ collapse, shock, and eventually death36. Another report by Liu and Li37 suggests that the binding of viral proteins attacks the heme localized on the hemoglobin 1-beta chain to detach the iron and to constitute the porphyrin. The above activity of viral protein attenuates the hemoglobin function to transport oxygen and CO2. This also interferes with the heme anabolic pathways of humans resulting in the several debilitating manifestations of COVID-1937. A review of SARS-CoV-2’s activity and its impact on the host’s immune system is discussed in detail in subsequent sections.
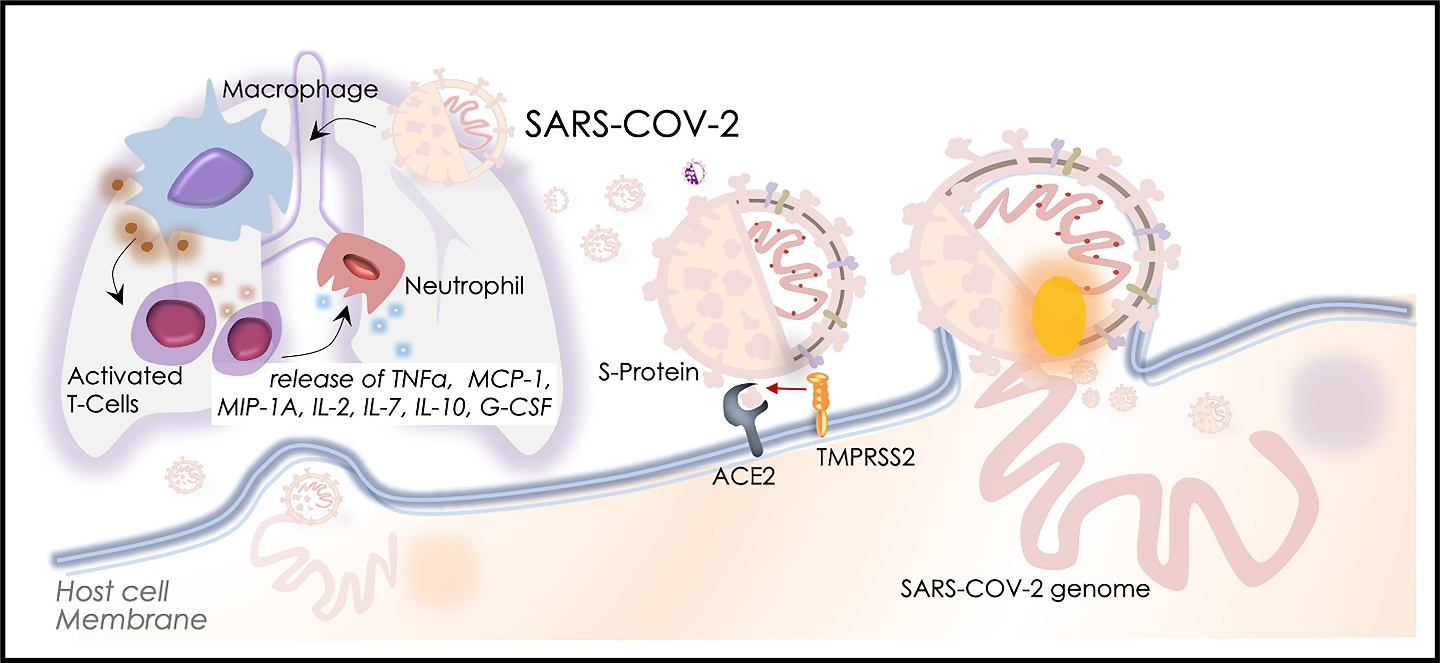
Figure 2: SARS-COV-2 Pathogenesis and immunological triggers. Schematic diagram showing SARS-COV-2 induced immunological trigger which involves its interaction with alveolar macrophages and subsequent activation of T-cells and neutrophils. The right side of the figure depicts SARS-COV-2 entry and release of its ssRNA genome into the host cells.
SARS-CoV-2 Infection and Immune Response
Innate immunity plays a crucial role in combating the severity of any viral infection. In a recent study, the innate immune response was shown to cause an increase in the neutrophil count and cause lymphopenia38 which indicates its critical role in disease progression and severity. The Severe Acute Respiratory Syndrome caused by contagious SARS-CoV-2 primarily targets the lungs and causes severe clinical manifestations. The frontline complement proteins, type I Interferons (IFN), and other factors of innate immune response essentially control viral replication in the host during the initial stages of the disease itself39. Activation of these factors on viral exposure subsequently triggers and shapes the adaptive immune response. However, an over-activation of innate immune response can result in lethality to host tissues40.
Previously, it has been shown that SARS-CoV attacks T cells. A minimal expression of ACE2 receptor on monocytes/macrophages in the lungs was observed by Dandekar and Perlman41, which suggests a possibility of the involvement of other immune mechanisms such as antigen-antibody reactions. It is speculated that SARS-CoV-2 may modulate the host innate immune response by dampening the type I IFN response as is often the case in SARS. However, as of now, there is no evidence which indicates a similar mechanism of modulation of host innate immune responses by SARS-CoV-242.
A lot has been learnt regarding the molecular pathogenesis and host immune responses of human coronaviruses from the SARS-CoV epidemic in 2003. Many studies were aimed at understanding the immune response in SARS-CoV infected patients so as to highlight upon the pathogenesis and clinical perseverance of the infection. These efforts to decipher the underlying immune mechanisms have investigated the interaction of SARS-CoV with innate immune cells e.g., dendritic cells (DC), macrophages, and also, their role in triggering other immune cells and responses including phenotype maturation, T- cell priming activity, cytokine production, and phagocytosis. Of note, the activated macrophages and DCs essentially contribute to the maintenance of innate response homeostasis within the host’s lung and inside infected tissues during the course of the infection. During the early phase of the COVID-19 pandemic, the pathology of SARS-Co-V-2 was still under investigation, and its molecular pathogenesis and function in host immune response remained largely uncharacterized, and as a result, knowledge of these aspects was majorly derived from the studies that investigated SARS-CoV immunopathogenesis (Fig. 3).
SARS-CoV-2 infections are characterized with a high degree of lymphopenia, which in some cases has been reported to affect CD4+ T cells, CD8+ T cells, B cells and natural killer cells, while other studies indicate that it is more associated with CD8+ T cells. Severe cases of COVID-19 also show a marked increase in the levels of cytokines such as IL-6, IL-10 or tumor necrosis factor (TNF), which are known to have a direct effect on T cells as well as DCs and neutrophils. T cell depletion may also be a result of hyperactivation or even the excessive expression of pro-apoptotic factors such as FAS, TRAIL or caspase 343.
The CD8+ T cell response in COVID-19 has been observed to vary within and among cohorts as well. Terminally differentiated or exhausted T cells have been associated with certain cases of severe COVID-19 owing either to an increased expression of the inhibitory receptors PD1, TIM3, LAG3, CTLA4, NKG2A and CD39, or as a result of hyperactivation, or in some cases, a result of decrease in the expression of inhibitory receptors has also been observed. In comparison with CD8+ T cell activation, CD4+ T cell activation is lesser. However, CD4+ responses are associated with worse clinical outcome in patients. SARS-CoV-2 S protein specific CD4+ T cells have a TH1 cell profile, and they have the ability to produce IFNÉ£, while the role of TH2 cells in COVID-19 is less clearly understood as of yet43.
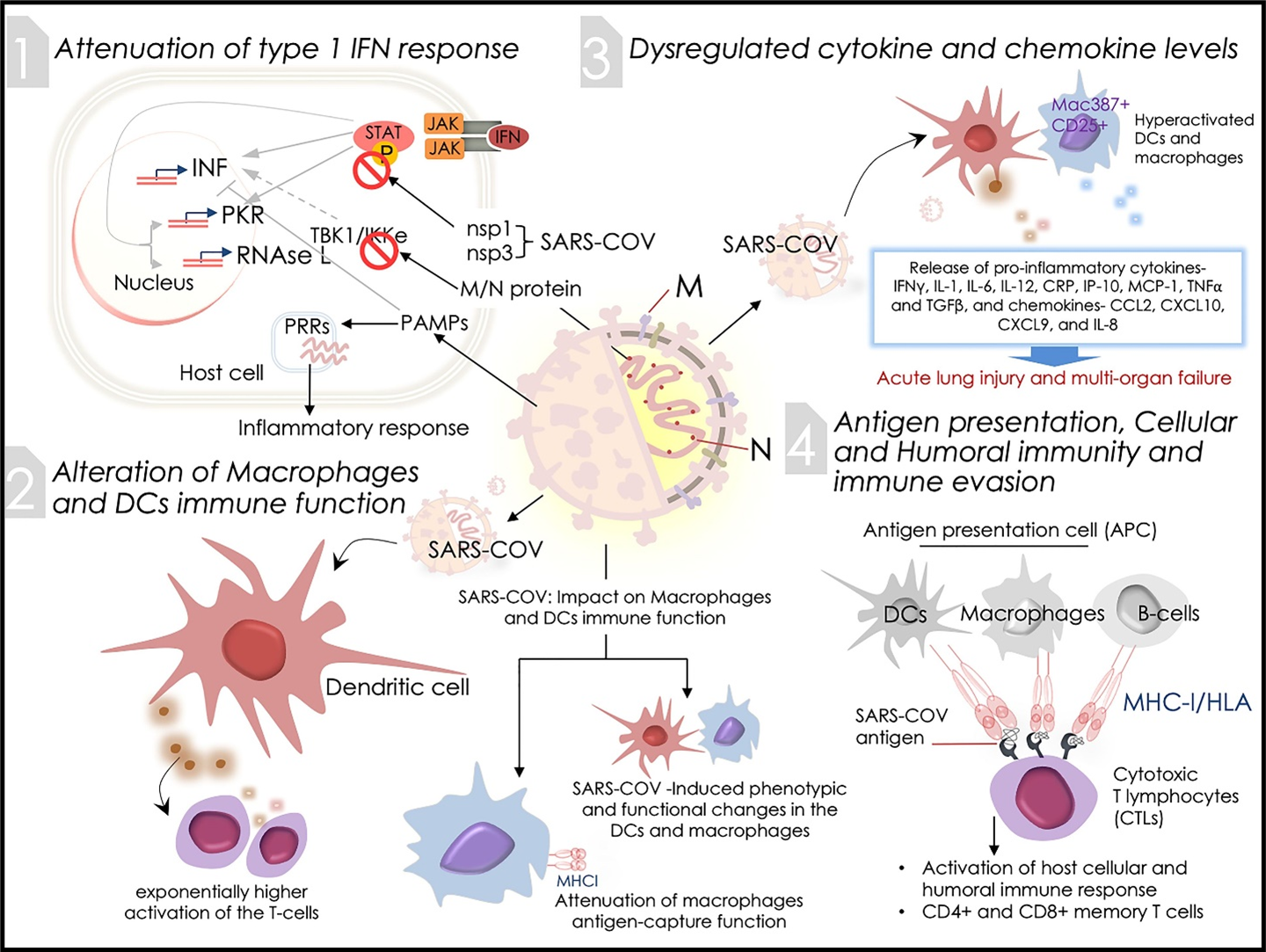
Figure 3: SARS-CoV-2 Infection and the host immune response. Schematic diagram showing immunomodulatory activities of coronavirus infection that includes its impact on (1) attenuation of type 1 IFN response, (2) alteration of macrophages and DCs immune function, (3) dysregulated cytokine and chemokine levels, and (4) antigen presentation, humoral immunity and immune evasion. Pictorials are inspired by the reference information obtained from SARS-CoV infection.
SARS-CoV-2 and Innate Immune Response
Attenuation of type 1 IFN response
Host interferons (IFN) are well-known to interfere with viral life cycle and limit their replication by activating several hosts antiviral effectors proteins such as Protein kinase R (PKR), RNAse L and other factors of innate response, but prior studies on SARS-CoV encoded proteins have been described to attenuate the IFN mediated antiviral signalling by inhibiting the IFN activity (Fig. 3)44. In the early phase of the SARS-CoV-2 pandemic, these insights on coronaviruses in general, included the SARS- and MERS-CoVs is what provided basic conceptual as well as speculative knowledge regarding the novel virus. The SARS-CoV nsp1 protein inhibits IFN function by modulating STAT phosphorylation, and a similar control over IFN activity has been observed by nsp3 involving unknown mechanisms. The IFN response blockage by SARS-CoV membrane (M) and nucleocapsid (N) structural proteins has also been shown to be mediated by TBK1/IKK through an unknown mechanism. Of note, attenuation of IFN production and/or function by the SARS-CoV structural and non-structural proteins activates inflammatory signalling, essentially by the progressive viral replication in host cells induced by Pathogen-Associated Molecular Patterns (PAMPs). The PAMP machinery in turn attenuates IFN function and elicits pattern recognition receptors (PRRs) promoting an inflammatory response. Deficiency of IFN response elevates counts of Ly6C low monocytes and neutrophils18,45 (Fig. 3). These aspects of SARS CoV pathogenesis and immune evasion were speculated to apply to SARS-CoV-2 as well.
With increasing research on the novel SARS-CoV-2, a better understanding of its immune mechanisms has been obtained. Being an RNA virus, upon cellular entry, several of the SARS-CoV-2 proteins have the ability to interact with and bind to RNA, even that of host origin. Banerjee et al. shed light upon the molecular basis of SARS-CoV-2 pathogenesis describe the ability of SARS-CoV-2 proteins to interact with human RNAs. They identified a very specific interaction between the viral NSP16 protein- which localizes in the nucleus upon infection, and the U1 and U2 snRNAs which mediate post-transcriptional RNA splicing, and in doing so, NSP16 has the ability to suppress global mRNA splicing in SARS-CoV-2 infected cells so as to decrease host protein and mRNA levels by triggering nonsense-mediated decay of improperly spliced mRNAs. This mechanism may result in inhibition of IFN-mediated immune responses since most of the key genes for IFN stimulation are spliced post-transcriptionally. Additionally, the SARS CoV-2 NSP1 binds to 18S ribosomal RNA in the mRNA entry channel of the 40S subunit and disrupts translation of RNA with an ability to suppress translation of global host mRNAs and the host IFN immune response. Similarly, NSP8 and NSP9 bind to the 7SL RNA component of the SRP (signal recognition peptide) preventing its proper folding and assembly which further suppresses the integration of integral membrane proteins in the cell membrane. A very interesting finding by Banerjee et al. was that these four viral proteins are all products of the initial stages of the virus replication cycle prior to production of the viral dsRNA which is a trigger for the type I IFN response, and this disruption of cellular processes is what allows the virus to replicate with a minimization in the IFN-mediated immune response46.
Additionally, NSP6 interferes with IFN-I production by binding TANK binding kinase 1 (TBK1) to suppress IRF3 phosphorylation. NSP13 and NSP15 bind and block TBK1 phosphorylation. The ORF6 localizes at the nuclear pore complex (NPC), binds importin karyopherin-α2 (KPNA2) to target the nuclear import pathway and inhibit the IRF3 nuclear translocation. Several SARS-CoV-2 proteins also have the ability to block STAT1/STAT2 phosphorylation. Nsp1, nsp6, nsp13, ORF3a, M, and ORF7b can inhibit STAT1 phosphorylation and ORF6 blocks STAT1 nuclear translocation, while nsp6, nsp13, ORF7a, and ORF7b prevent STAT2 phosphorylation47.
SARS-CoV-2 and altered Macrophages and Dendritic cell immune function
Neither DCs nor MP cells were shown to acquire SARS-CoV infection, or even facilitate viral replication. However, reports have shown that exposure to the infectious SARS-CoV acquires phenotypic and functional changes in the DC and macrophages (Fig. 3). These changes alter differentiation of the DCs and modulate the expression of co-stimulatory molecules required for priming of T-cells, which serves as an important factor for stimulating the subsequent adaptive immune response. Exposure of infectious SARS-CoV was earlier shown to attenuate the antigen-capture function of macrophages, wherein endocytosis uptake gets disrupted. This, in turn, substantially compromises the antigen presentation function of macrophages to T-cells. On the other hand, exposure of differentiated DCs to infectious SARS-CoV results in exponentially higher activation of the T-cells by these cells. These results offers insights into the immunomodulatory function of SARS-CoV in the innate immune cells that further determine the adaptive response in the event of SARS-CoV infections (Fig. 3)18, 48, 49.
In the case of SARS-CoV-2, alveolar macrophages are most likely the very first immune cells that are encountered by the virus during infection. A study by Dalskov et al. indicates that naïve alveolar macrophages are incapable of sensing SARS-CoV-2 and mounting an IFN response against the virus, which suggests that SARS-CoV-2 genomic RNA is not detected by alveolar macrophages and this is a postulated reason for the early asymptomatic phase of COVID-19. This is very much in contrast to the response mounted by alveolar macrophages against other respiratory viruses such as the Influenza virus and the Sendei virus. However, Dalskov et al. noted that similar to SARS and MERS, SARS-CoV-2 was incapable of causing productive infections in alveolar macrophages50.
Dysregulated cytokine and chemokine levels and its clinical consequences
The attenuated type 1 IFN action, as discussed earlier, compromises early viral control. Additionally, the SARS-CoV replication rate in ACE+/+ human lung epithelial cells are very rapid. Therefore, these factors potentially result in hyperactivation of dendritic cells and macrophages, aggravating the proinflammatory responses of these innate immune cells (Fig. 3). Additionally, elevated expressions of IFN-γ, IL-1, IL-6, IL-12, and TGFβ pro-inflammatory cytokines and CCL2, CXCL10, CXCL9, and IL-8 chemokine were observed in the blood serum of severely-sick SARS-CoV patients as compared to patients having moderate symptoms (Fig. 3)51, 52. Studies have shown a significant increase in levels of pro-inflammatory cytokines (PICs) like IL-2, IL-7, CRP, IP-10, MCP-1, TNF-alpha – all further validating the presence of a modulated, hyperactive innate immune response in SARS-CoV infected patients53-55. A clinical outcome of the release of PICs in SARS-CoV-infected ACE2+ cells in patient tissues was suggested to promote acute lung injury and multi-organ failure (Fig. 3). For instance, MCP-1 functions in the induction of the migration of macrophages/monocytes and potentiate these cells in the vascular system to travel to the infested area/tissue to ensure their combative function against the viral agent(s). However, the combative function of these cells is not limited to infected cells only, thus it can also harm normal cells that are in close affinity. In severe SARS-CoV cases, it has been consistently found that there is a higher infiltration of monocyte-macrophages-neutrophils at the site of infection. This provides a framework for understanding the pathogenesis of SARS-CoV infection. Some PICs e.g., TNF-α, TGF-β1, etc. producing macrophages undergo proliferation and become activated, resulting in a higher PICs release, causing more damage of the host tissues56. Expression of Mac387+ and CD25+ proliferation and/or activation markers on a large number of alveolar macrophages corroborates these results. However, TGF-β1 has been implicated in alveolar epithelial cell death and this may promote Fas-mediated cell death of platelets and lymphocytes as well. These TGF-β1 function may produce acute lung injury, thrombocytopenia, and lymphopenia in SARS-CoV patients. Expression levels of apoptotic Fas and Fas Ligand (FasL) remain high in pneumocytes and epithelial cells of the lungs of SARS patients53,56,57.
SARS-CoV-2 infection and Antigen Presentation
Antigen presenting cells (APCs) are the host’s major anti-viral defense mechanism that process and present the antigen when pathogens strike into cells. The major histocompatibility complex (MHC) or human leukocyte antigen (HLA) presents the antigenic peptides that are primarily recognized by cytotoxic T lymphocytes (CTLs) (Fig. 3). Although, the process of antigen presentation remains highly conserved and demonstrates mechanistic uniformity for diverse viral infections, lack of any evidence of these mechanisms in SARS-CoV-2 presently hindered a clear view. The gathered knowledge on SARS-CoV and MERS-CoV for their antigen presentation mechanisms offers an insight into these functions for SARS-CoV-2 (Fig. 3). The coronavirus antigen presentation is mostly governed by MHC I molecules58, but then MHC II similarly subsidizes to antigen presentation.
Early reports have shown that several HLA allelic polymorphisms including HLA-DR B1*1202, HLA-B*0703, HLA-B*4601, and HLA-Cw*0801 are associated with vulnerability to SARS-CoV,59 however, three alleles viz. the HLA-A*0201, HLA-DR0301, and HLA-Cw1502 are also associated with host’s defense mechanism against the SARS-CoV infection60. Reports examining the MHC II polymorphisms in MERS-CoV infection revealed that antigen presented by two MHC II molecules viz. HLA-DQB1*02:0 and HLA-DRB1*11:01 contributes to the vulnerability to MERS-CoV61. Moreover, polymorphisms of antigen presentation related mannose-binding lectin (MBL) gene were found to be predictive of a greater risk of SARS-CoV disease62.
SARS-CoV-2 and Cellular and Humoral Immunity
Antigen presentation consequently activates the cellular and humoral immune response in the host, which is primarily triggered by viruses targeting B- and T- cells. The immunoreactive antibodies against SARS-CoV show a characteristic IgM pattern that disappears at the end of the 12th week, while IgG antibodies stay in the humoral system for a long time63. The IgG antibodies showing specificity to SARS-CoV mainly includes the S- and N-specific antigen64. A recent report showed the counts of CD4+ and CD8+ T lymphocytes significantly reduce in the blood along with the peripheral blood mononuclear cells (PBMCs) in COVID-19 patients, however, these cells showed a hyper-activated state as marked by high expression of HLA-DR (CD4+) and CD38 (CD8+) dual positive populations65. Severely infected SARS-CoV patients were also earlier shown to have a depletion of CD4+ and CD8+ T cells population. Of note, in the survived SARS-CoV patients, the CD4+ and CD8+ memory T cells population was present for about four years and has actively acquired DTH response, proliferation, and IFN-g production66. A study conducted by Tang et al.67, showed that memory T cells retain SARS-CoV S-peptide specific memory even after six years of the viral infection that was found to be evident in 14 out of 23 patients. Similarly, a potent activity of CD8+ T cells was also validated in the mice model for the clearance of MERS-CoV viral load68. These evidences could provide a valuable reference to assess the host’s immune response against the SARS-CoV-2 in current clinical settings (Fig. 3).
SARS-CoV-2 Immunogenicity and Cytokine Storm
Development of ARDS largely contributes to the mortality in COVID-19 patients. Huang et al. have shown that 6 out of 41 SARS-CoV-2 patients died of ARDS18. ARDS is a frequent immunopathological response as is evidently seen in consecutive coronavirus infections65. An activated systemic inflammatory response in COVID-19 patients prompts an array of pro-inflammatory chemokines and cytokines that contributes to the emergence of a robust cytokine storm in the patients. It includes the constitutive release of IFN-a, IFN-g, TNF-a, IL-1b, IL-6, IL-12, IL-18, IL-33, and TGF-b pro-inflammatory cytokines and CXCL9, CXCL10, CCL2, CCL3, CCL5, CXCL8, chemokines (Fig. 3)18,45,69,70. Consistent to these findings in COVID-19, patients with severe MERS-CoV infections exhibited a higher level of pro-inflammatory cytokines and chemokines in the serum, in comparison to those who had moderate infection71. The clinical feature of cytokine storm involves aggressive assault of the immune system in the host body that leads to ARDS and multiple organ failures and eventually contributes to mortality in severely infected SARS-CoV2 patients, as also reported earlier in MERS and SARS-CoV pathologies (Fig. 3)65.
SARS-CoV-2 and Immune Evasion
MERS and SARS coronaviruses evade the host immune system to promote their replication and survival. Microbe-derived molecules/motifs viz. PAMPs (Pathogen-associated molecular pattern) are largely recognized by pattern recognition receptors (PRRs) or toll-like receptors (TLRs) that further affirms the response of the host’s immune system. MERS and SARS coronaviruses were shown to instigate the formation of double-membrane vesicles, lacking PRR that facilitates their replication in the vesicles and escaping from the host identification of their dsRNA. Interferon-I (IFN-I) provides protection to host against viral infection, however, inhibition of its above function was observed in MERS-CoV and SARS-CoV infected mice72,73. The role of MERS-CoV accessory protein 4a was suggested earlier to inhibit IFN activation74. In a similar manner, MERS-CoV virus membrane antigens ORF4a, ORF4b, and ORF5 were seen to restrict IFN regulatory factor 3 (IRF3) nuclear localization that subsequently impairs IFN promoter activation75. These findings emphasized the importance of building preventive/therapeutic strategies that could target the immune evasion function of SARS-CoV-2 (Fig. 3).
COVID-19 Diagnostics: Clinical and Laboratory Approaches
In clinical diagnosis, patient history is designated as an essential primary record. Prior to an advancement in the stage of this pandemic and the diagnosis of infection, patient history, in terms of contact with suspected or confirmed COVID-19 cases, supported quicker diagnosis for COVID-19 (Fig. 4). At the physiological level, mild-moderate fever, coughing/sore throat, and shortening of breath are primary symptoms of SARS-CoV-2 infection patient (Fig. 4)76. Nevertheless, reports have shown that SARS-CoV-2 may persist in asymptomatic people for more than two weeks and there are chances of its recurrence after recovery as well77. The infected individual, at an early stage, may possess an average or reduced count of peripheral white blood cells (WBCs). A recent clinical study, comprising a 1,099 COVID-19 patient’s cohort exhibited a lesser than 4×109/L WBC count78. Also, most of the COVID-19 patients were found to have elevated enzymes, proteins like C-reactive protein (CRP), aspartate aminotransferase (AST), myoglobin, and erythrocyte sedimentation rate (Fig. 4)74,78,79. As a preferred clinical diagnosis method, analyses of chest X-ray of a majority of COVID-19 patients has revealed an abnormal state of the lungs, marked by shadows in small patches and interstitial changes that are excessively evident in the extra-pulmonary bands80. Furthermore, Computed Tomography (CT) of the chest in COVID-19 patients showed several extended ground-glass opacities and infiltrations with the progression of infection’s severity (Fig. 4)81. Often, patients with COVID-19 infection exhibit acute lung injury, unusual pneumonia, and ARDS36. At the advanced stage, increased inflammation, accumulation of fluid, and advanced fibrosis in the COVID-19 patient’s lungs was shown to compromise the gas exchange and thus may postulate an eventual organ failure (Fig. 4)82. Therefore, a greater abnormality in chest radiographic scans could predict a more severity of the COVID-19 infection in the patient.
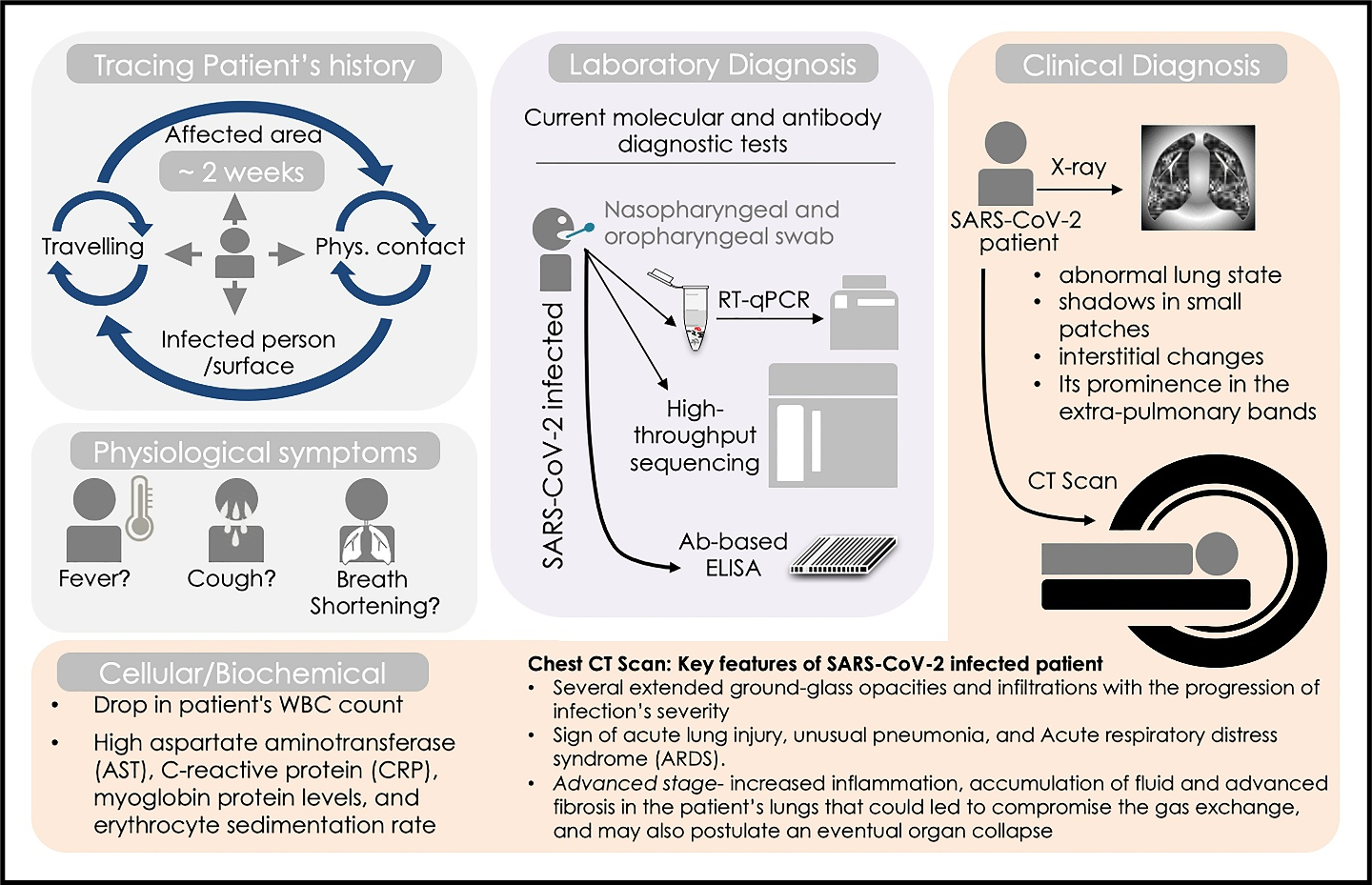
Figure 4: COVID-19 diagnostics: Clinical and laboratory approaches. Schematic diagram showing earliest diagnosis step including consulting the SARS-COV-2 suspects travel and contact history, physical examination for fever, coughing, and sign of shortening of breath. Laboratory diagnosis approaches include RT-qPCR and high-throughput sequencing analyses for SARS-COV-2 nucleic acids in samples collected from patent’s nasopharyngeal and oropharyngeal swab. Clinical diagnostic regime includes chest X-ray, CT scan and analyses of patent’s cellular/biochemical parameters.
Quantification of COVID-19 nucleic acid in nasopharyngeal and oropharyngeal swab sample or secretion of the respiratory tract sample by quantitative Reverse Transcription- Polymerase Chain Reaction (RT-qPCR) is the routine diagnostic approach enrolled presently in the laboratories worldwide (Fig. 4)83,84. RT-qPCR remains the gold standard and sensitive tool for COVID-19 detection in comparison to other available symptomatic and scanning tests as it directly detects the SARS-CoV-2 genetic signature (RNA) and quantifies the viral load. Therefore, RT-qPCR offers enormous help to clinicians and epidemiologists to trace COVID-19 spread either to individual clusters or in the community, and for its clinical management as well85. The RT-qPCR test includes primer and probe designing at initial steps followed by the optimization of the amplification conditions, while modification in processing steps and duration can be conceived based on manufacturer’s recommendation. Primers and probe for RdRP (RNA-dependent RNA polymerase) gene of the ORF1ab region of open reading frame and E (Envelope -protein) gene have shown high analytical sensitivity85-87. As an early readout of RT-qPCR utility, initial reports from China revealed that its overall detection rate for SARS-CoV-2 infection was 38% in 4880 cases88, while other testing performed on 51 confirmed patients of SARS-CoV-2 infection showed 71% positive detection rate89. It has been observed that patients may show COVID-19 positive in the RT-qPCR test after 2- 8 days of getting infected84,90. Some modified methods of RT-qPCR test known as Reverse Transcription Loop-mediated isothermal amplification (RT-LAMP) and CRISPR–Cas12-based assay for SARS-CoV-2 RNA detection were also developed and enrolled for diagnosis91,92. Consistent to these novel diagnostic initiatives and with evolving situation in the United States, the FDA (Food and Drug Administration, USA) has recently approved the emergency usage authorization of cobas 6800/8800 high throughput platforms for SARS-CoV-2 diagnosis which offer quality and high-volume testing capacity93, 94. High-throughput sequencing for SARS-CoV-2 nucleic acid and SARS-CoV-2 specific antibody-based ELISA systems have now been employed as point-of-care tests (POCT) for rapid SARS-CoV-2 diagnosis (Fig. 4)95.
Preventive and Therapeutic Strategies
Presently, we lack any SARS-CoV-2 -specific therapeutics that could offer a sure shot clinical outcome. Researchers and health professionals across the globe are summing up their efforts to identify and validate precise SARS-CoV-2 targeted therapeutics. Earlier, WHO had released technical guidance systematically listing the crucial measures to contain and further prevent COVID-19 spread96. It broadly specifies the strategic measures to prevent infection in healthcare professionals and related facilities and extensively discusses quarantine measures and requisite of rational use of masks and personal protective equipment (PPE)96. Hand-washing and usage of the sanitizer (containing > 60% of alcohol) or soap with water is well-recommended to effectively prevent COVID-1997. It is also advised that keeping the social distancing of at least 1-meter between COVID-19 patients and an uninfected person can significantly mitigate its rate of transmission98.
By date, we lack an FDA approved antiviral to cure COVID-19 patients or prevent SARS-CoV-2 infections. Presently, the supportive care including supplementary oxygen, mechanical ventilation, infection prevention, and control measures are broadly being utilized for clinical management of COVID-19. Several clinical trials are now underway and being regularly updated on USA (https://clinicaltrials.gov/ct2/results?cond=COVID-19), EU (https://www.clinicaltrialsregister.eu/ctr-search/search?query=covid-19), and at the WHO’s International Clinical Trials Registry Platform (ICTRP; available at https://www.who.int/ictrp/en/) online portals. The alternative therapeutic options that are presently being envisioned to cure COVID-19 include RNA vaccine, oligonucleotide-based therapies, monoclonal antibodies, plasma, and interferon-based therapies25,99,100. The progress on vaccine development has already surpassed several stages of trials and vaccinologists are now assessing the immune-protection against COVID-19 at the National Institutes of Health, USA, China, and UK96,101. The broad-spectrum antiviral drugs like HIV protease inhibitors and nucleoside were repurposed and are also being utilized as therapeutic alternatives given their benefits against other earlier reported 2 viral infections100,102. As of recent data, more than 85 % percent of patients in China received various anti-viral agents including oseltamivir, ganciclovir, and lopinavir/ritonavir15. Another study showed that repurposing of remdesivir could help in COVID-19 treatment, and this drug has been approved for the same103. Additionally, a recent report discussed benefits of early-stage intravenous glucocorticoid administration (methylprednisolone, 40–80 mg, given once a day till 5 days), while its treatment course can be modified based on clinician’s recommendation104. Furthermore, immune suppressants were recommended to all patients undergoing diagnosis of accompanying hyper-inflammation. Another insightful report citing readouts of multi-centric clinical trials in China demonstrated that chloroquine phosphate i.e., a drug used for malaria treatment, exhibits a clear efficacy and protection against COVID-19-associated pneumonia105. FDA’s recent approval to use convalescent plasma (collected from COVID-19 exposed patients) also gave a remedial hope to critically ill COVID-19 patents106. Presently, a few potential therapies have been developed for treatment, one such example is the IL-6 receptor blockade. In this line, China has initiated multi-centric, randomized controlled trials to use tocilizumab for COVID-19 patients who are having pneumonia and elevated IL-6 levels107. Other therapeutic options include the usage of specific cytokine blockade by tocilizumab or anakinra and through JAK inhibition (Available at https://www.clinicaltrials.gov/ct2/show/NCT04322773; accessed on 16 April 2020). Currently, unprecedented research efforts towards finding the treatment possibilities for COVID-19 have been made and key recommendations are constantly updated worldwide. Evidence-based details of recent potential therapeutic strategies for COVID-19 cure are duly enlisted in Table 1 and details of vaccines and technology used are duly listed in table 2.
Table 1: Evidence-based details of recent and priory utilized and available potential therapeutic possibilities to cure COVID-19.
|
Repurposing of FDA approved drug |
Type of Drug |
Drug or Molecule |
Current Use |
Mechanism and Current status |
References |
|
Anti-malarial agent |
Chloroquine Phosphate & Hydroxychloroquine sulfate |
Malaria Treatment |
- Anti-malarial agent - Effective against viral entry & killing through photolytic processing - Not recommended for COVID-19 therapy any longer. |
[14,108] |
|
|
Anti-Viral Agents |
Remdesivir |
Ebola Virus Treatment |
- Nucleotide analogue - Approved for COVID-19 therapy |
[109] |
|
|
Ritonavir &Lopinavir |
HIV Treatment |
- Protease inhibitor - Shown in vitro activity against other coronaviruses - No treatment benefit observed in initial randomized, controlled, open-label trial for COVID-19 patients |
[100,102,110] |
||
|
Favipiravir (T-705) |
Influenza treatment
|
- RNA polymerase inhibitor thus inhibits virus replication - Has broad spectrum anti-viral activity - Tested in clinical trial |
[100,111] |
||
|
Ribavirin |
Treatment of Respiratory |
- Inhibits viral RNA-dependent RNA - Polymerase - Used in the treatment of other coronaviruses - Under screening and development |
[109,112] |
||
|
Umifenovir (Arbidol) |
Influenza Treatment |
- Inhibits viral S Protein & ACE2 interaction - Repurposed for COVID treatment and being tested in clinical trial |
[113] |
||
|
Antibodies |
Tocilizumab (Actemra) |
Treatment of Rheumatoid Arthritis |
- Monoclonal antibody for IL-6 receptor - Under testing in clinical trial |
[114] |
|
|
Antibody CR3022 |
Neutralizing Antibody |
- Neutralizing Ab, target- viral (SARS-CoV) receptor-binding domain (RBD) isolated from convalescent plasma of COVID-19 patients |
[115] |
||
|
Interferon |
IFN-α2b |
Hepatitis C |
- Type I interferon - Tested in clinical trial |
[111] |
|
|
Future Drug Targets |
Proteins |
Non-structural proteins (nsp10/nsp16) |
Under development |
- Inhibitor of Viral Scaffolding and RNA synthesis - Under screening and development |
[116] |
|
mRNA vaccine |
Anti-viral vaccine (mRNA-1273) |
Authorized for use |
- Messenger RNA (mRNA) - Authorized for use |
[99,101] |
|
|
Herbal |
Herbal treatments |
Influenza A treatment SARS-CoV treatment |
- Chinese traditional medicine - Under screening and development |
[117, 118] |
|
|
Antibodies |
Monoclonal antibody therapy
Convalescent sera from priorly infected patients |
SARS-CoV treatment
COVID-19 therapy |
- Anti-SARS-CoV-2 &anti-ACE2 monoclonal antibodies - Under screening and development - Currently being used for treatment. |
[25,99] |
|
|
Enzymes |
Soluble angiotensin-converting enzyme 2 |
Under development |
- Soluble recombinant ACE2 protein - Under screening and development |
[119] |
#Other potential sources of information are-
- https://clinicaltrials.gov
- https://www.who.int/ictrp/en/
- https://www.gilead.com/purpose/advancing-global-health/covid-19/remdesivir-clinical-trials
- WHO landscape analysis of therapeutics for coronavirus as on 21st March 2020
Table 2. Evidence-based details of recent and priory utilized and available potential therapeutic possibilities to cure COVID-19.
|
S. No. |
Vaccine |
Vaccine developer |
Technology |
|
1. |
(Ad5-nCoV Convidicea) |
CanSino Biologics, Institute of Biotechnology of the Academy of Military Medical Sciences, China |
Adenovirus vector vaccine (recombinant adenovirus type 5 vector) |
|
2. |
BBIBP-CorV |
Sino pharma Institute of Biological Products, Wuhan Institute of Biological Products, China |
SARS-CoV-2 vaccine (vero cells) |
|
3. |
BBV152 (Covaxin) |
Bharat Biotech, Indian Council of Medical Research, India |
Inactivated SARS-CoV-2 vaccine |
|
4. |
Sinovac, China |
Inactivated SARS-CoV-2 vaccine |
|
|
5. |
CoviVac |
The Chumakov Centre at Russian Academy of Sciences, Russia |
Inactivated SARS-CoV-2 vaccine |
|
6. |
EpiVacCorona |
Vector Institute, Russia |
|
|
7. |
(Johnson & Johnson), BIDMC, Netherlands, United States |
Adenovirus vector vaccine (adenovirus serotype 26) |
|
|
8. |
University of Oxford, AstraZeneca, CEPI, United kingdom |
Adenovirus vector vaccine (modified chimpanzee adenovirus vector, ChAdOx1) |
|
|
9. |
Pfizer–BioNTech United States, Germany |
RNA vaccine (modRNA encapsulated in lipid nanoparticles) |
|
|
10. |
Gamaleya Research Institute of Epidemiology and Microbiology, Russia |
Adenovirus vector vaccine (recombinant adenovirus type 5 and 26 vector) |
|
|
11. |
ZyCoV-D |
Cadila Healthcare, India |
DNA vaccine (plasmid expressing SARS-CoV-2 S protein) |
|
12. |
Virus-like particles[f] (recombinant, plant-based with AS03) |
Conclusion
In the present review, we have provided insights into the SARS-CoV-2 pathology by shedding light on the host’s immune function comprising innate, cellular, and humoral responses. Further, we discussed the immunomodulatory functions of SARS-CoV-2 including cytokine storm and immune evasion. The review further sheds light on the clinical and laboratory strategies that are presently employed for COVID-19 diagnosis. We herein provide a detailed and up-to-date insight into the preventive and mitigating measures that are presently being considered against COVID-19 globally. The updates on the repurposing of FDA approved drugs to cure COVID-19 have been highlighted and reflect state of ongoing research and therapeutic initiatives those are being employed worldwide towards finding the treatment strategies for COVID-19. Furthermore, it conferred an update on revised diagnostic state and strategies for accelerated detection of COVID-19 infection. The ongoing clinical trials and research worldwide to develop the interventional COVID-19 treatment have been specifically envisaged. Given the fact that being a scientific community, we are now engaged in translating the health implications of ongoing COVID-19 pandemic, an extensive research further is required to address the challenges of COVID-19 pandemic with identifying the druggable targets, vaccine development, quicker diagnosis, overcoming the acute immune-response linked with SARS-CoV-2 infection.
Declaration of competing interest
The authors declare no conflicts of interest.
Funding
Nil
References
- Wu Y, Ho W, Huang Y, et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 2020; 395: 949-950.
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 2016; 3: 237-261.
- Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil Med Res 2020; 7: 1-10.
- Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Society Exp Biol Med 1966; 121: 190-193.
- Holmes KV. SARS coronavirus: a new challenge for prevention and therapy. J Clin Invest 2003; 111: 1605-1609.
- Skowronski DM, Astell C, Brunham RC, et al. Severe acute respiratory syndrome (SARS): a year in review. Annu Rev Med 2005; 56: 357-381.
- Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015; 386: 995-1007.
- JandriÄ P. Postdigital Research in the Time of Covid-19. Postdigit Sci Edu 2020;2: 233-238.
- Lu H, Stratton CW, Tang YW. Outbreak of Pneumonia of Unknown Etiology in Wuhan China: the Mystery and the Miracle. J Medl Virol 2020; 92: 401–402.
- Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395: 507-513.
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395(10224): 565-74.
- Hui DS, Azhar EI, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J of Infect Dis 2020; 91: 264.
- Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 2020; 2020: 102433.
- Pang J, Wang MX, Ang IYH, et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel Coronavirus (2019-ncoV): a systematic review. J Clin Med 2020; 9: 623.
- Lai CC, Shih TP, Ko WC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 2020; 2020: 105924.
- Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020; 172: 577-582.
- Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 2020; 382: 1199-1207.
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497-506.
- Yang J, Zheng Y, Gou X, et al. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: a systematic review and meta-analysis. Int J Infect Dis 2020; 94: 91-95.
- Adhikari SP, Meng S, Wu YJ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty 2020; 9: 1-12.
- Fauci AS, Lane HC, Redfield RR. Covid-19—Navigating the Uncharted. Mass Medical Soc; 2020.N Engl J Med 2020; 382: 1268-1269.
- Cai J, Sun W, Huang J, et al. Indirect Virus Transmission in Cluster of COVID-19 Cases, Wenzhou, China, 2020. Emerg Infect Dis 2020; 26: 1343-1345.
- van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Eng J Med 2020; 382: 1564-1567.
- Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J Microbiol Immunol Infect 2020. doi.org/10.1016/j.jmii.2020.03.022
- Shanmugaraj B, Siriwattananon K, Wangkanont K, et al. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac J Allergy Immunol 2020; 38: 10-18.
- Zhang H, Penninger JM, Li Y, et al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 2020; 46: 586-590.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271-280.
- Reinke LM, Spiegel M, Plegge T, et al. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS One 2017; 12: e0179177
- Deshotels MR, Xia H, Sriramula S, et al. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin ii type i receptor–dependent mechanism. Hypertension 2014; 64: 1368-1375.
- Shah PP, Wang T, Kaletsky RL, et al. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol Pharma 2010; 78: 319-324.
- Mingo RM, Simmons JA, Shoemaker CJ, et al. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J Virol 2015; 89: 2931-2943.
- Wang H, Yang P, Liu K, et al. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res 2008; 18: 290-301.
- Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARSâCoVâ2. Protein Sci 2020. doi.org/10.1002/pro.3873.
- Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020; 395: 514-523.
- Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol Sin 2020. Preprint.
- Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 2020; 323: 1061-1069.
- Liu W, Li H. COVID-19: Attacks the 1-Beta Chain of Hemoglobin and Captures the Porphyrin to Inhibit Human Heme Metabolism. ChemRxiv. 2020. Preprint.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273.
- Katze MG, He Y, Gale M. Viruses and interferon: a fight for supremacy. Nature Rev Immunol2002; 2: 675-687.
- García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science2006; 312: 879-882.
- Dandekar AA, Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nature Rev Immunol 2005; 5: 917-927.
- Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol2020; 38: 1-9.
- Chen Z & Wherry EJ. T cell responses in patients with COVID-19. Nature Reviews Immunology 2020; 20.
- Stark GR, Kerr IM, Williams BR, et al. How cells respond to interferons. Ann Rev Biochem 1998; 67: 227-264.
- Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. In Sem in Immunopath2017; 39: 529-539.
- Banerjee AK, Blanco MR, Bruce EA, et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 2020; 183(5): 1325-1339.
- Quarleri J, Delpino MV. Type I and III IFN-mediated antiviral actions counteracted by SARS-CoV-2 proteins and host inherited factors. Cytokine & Growth Factor Reviews 2021.
- Yilla M, Harcourt BH, Hickman CJ, et al. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res2005; 107: 93-101.
- Tseng CTK, Tseng J, Perrone L, et al. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J Virol2005; 79: 9470-9479.
- Dalskov L, Møhlenberg M, Thyrsted J, et al. SARS-CoV-2 evades immune detection in alveolar macrophages. EMBO reports 2020; 21(12): e51252.
- Cheung CY, Poon LL, Ng IH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol2005; 79: 7819-7826.
- Law HKW, Cheung CY, Ng HY, et al. Chemokine up-regulation in sars-coronavirus–infected, monocyte-derived human dendritic cells. Blood2005; 106: 2366-2374.
- Hayden FG, Fritz R, Lobo MC, et al. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest1998; 101: 643-649.
- Theron M, Huang KJ, Chen YW, et al. A probable role for IFN-γ in the development of a lung immunopathology in SARS. Cytokine2005; 32: 30-38.
- Jiang Y, Xu J, Zhou C, et al. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med2005; 171: 850-857.
- Cheung CY, Poon LLM, Lau AS, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 2002; 360: 1831-1837.
- Li H, Liu L, Zhang D, et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. The Lancet 2020; 395: 1517-1520.
- Liu J, Sun Y, Qi J, et al. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J Infect Dis2010; 202: 1171-1180.
- Keicho N, Itoyama S, Kashiwase K, et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Human Immunol2009; 70:527-531.
- Wang SF, Chen KH, Chen M, et al. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol 2011; 24: 421–426.
- Hajeer AH, Balkhy H, Johani S, et al. Association of human; leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Annals Thorac Med 2016; 11: 211.
- Tu X, Chong WP, Zhai Y, et al. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect2015; 71: 101-109.
- Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Eng J Med2003; 349: 508-509.
- de Wit E, van Doremalen N, Falzarano D, et al. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol2016; 14: 523.
- Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respirat Med2020; 8: 420-422.
- Fan YY, Huang ZT, Li L, et al. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Archives of virology 2009; 154(7): 1093-1099.
- Tang L, Wang L, Tan X, et al. Adenovirus serotype 7 associated with a severe lower respiratory tract disease outbreak in infants in Shaanxi Province, China. Virol J2011; 8: 23.
- Zhao J, Li K, Wohlford-Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Nat Acad Sci2014; 111: 4970-4975.
- Cameron MJ, Bermejo-Martin JF, Danesh A, et al. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res2008; 133: 13-19.
- Williams JL, Holman DW, Klein RS. Chemokines in the balance: maintenance of homeostasis and protection at CNS barriers. Front Cell Neurosci2014; 8: 154.
- Min CK, Cheon S, Ha NY, et al. Comparative and kinetic analysis of viral shedding and immunological responses in MERS patients representing a broad spectrum of disease severity. Sci Rep2016; 6: 1-12.
- Channappanavar R., Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016; 19: 181-193.
- Channappanavar R, Fehr AR, Zheng J, et al. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 2019; 129.
- Niemeyer D, Zillinger T, Muth D, et al. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol2013; 87: 12489-12495.
- Yang Y, Zhang L, Geng H, et al. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell2013; 4: 951-961.
- Wang Z, Yang B, Li Q, et al. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis 2020. doi.org/10.1093/cid/ciaa272.
- Yi Y, Lagniton PN, Ye S, et al. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci 2020; 16: 1753-1766.
- Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Eng J Med2020; 382: 1708-1720.
- Qiu H, Wu J, Hong L, et al. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis 2020; 20: 689-696.
- Jacobi A, Chung M, Bernheim A, et al. Portable chest X-ray in coronavirus disease-19 (COVID-19): A pictorial review. Clin Imag 2020; 64: 35-42.
- Li W, Cui H, Li K, et al. Chest computed tomography in children with COVID-19 respiratory infection. Ped Radiol 2020; 50: 796–799.
- Gralinski LE, Baric RS. Molecular pathology of emerging coronavirus infections. J Pathol 2015; 235: 185-195.
- World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance. 2020 (No. WHO/COVID-19/laboratory/2020.4).
- Zhai P, Ding Y, Wu X, Long J, Zhong Y, Li Y. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrobl Agents 2020; 105955.
- Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano2020; 14: 3822-3835.
- Corman V, Bleicker T, Brünink S, et al. Diagnostic detection of 2019-nCoV by real-time RT-PCR. World Health Organization. 2020.
- Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. 2020; 20: 453-454.
- Liu R, Han H, Liu F, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clinica Chimica Acta 2020; 505: 172-175.
- Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology 2020; 200432.
- Huang P, Liu T, Huang L, et al. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology2020; 295: 22-23.
- Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol 2020; 1-5.
- Yan C, Cui J, Huang L, et al Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 2020; 2020.
- Moran A, Beavis KG, Matushek SM, et al. The Detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 Assays. J Clin Microbiol 2020; 2020.
- Poljak M, Korva M, Gašper NK, et al. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol 2020.
- Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic Testing for Severe Acute Respiratory Syndrome–Related Coronavirus-2: A Narrative Review. Annals of Intern Med 2020.
- World Health Organization. Coronavirus disease (COVID-19) technical guidance: Infection prevention and control. 2020.
- Bauer-Savage J, Pittet D, Kim E, et al. Local production of WHO-recommended alcohol-based handrubs: feasibility, advantages, barriers and costs. Bulletin of the World Health Organization 2013; 91: 963-969.
- Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. Jama2020; 323: 1837-1838.
- Hodgson J. The pandemic pipeline. Nat Biotechnol. 2020. DOI: 10.1038/d41587-020-00005-z.
- Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) 2020; 19: 149-150.
- Liu C, Zhou Q, Li Y, et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci2020; 6: 315-331.
- Chu C, Cheng V, Hung I, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 2004; 59: 252-256.
- Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Eng J Med 2020; 382: 929-936.
- Zhou YH, Qin YY, Lu YQ, et al. Effectiveness of glucocorticoid therapy in patients with severe novel coronavirus pneumonia: protocol of a randomized controlled trial. Chin Med J 2020; 10.
- Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 2020; 14: 72-73.
- Tanne JH. Covid-19: FDA approves use of convalescent plasma to treat critically ill patients. BMJ2020; 368: 1256.
- Mehta P, McAuley DF, Brown M, et al. Across Speciality Collaboration, COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet2020; 395: 1033.
- Huang M, Tang T, Pang P, et al. Treating COVID-19 with chloroquine. J Mol Cell Biol2020; 12: 322-325.
- Al-Tawfiq JA, Al-Homoud AH, Memish ZA. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med Infect Dis 2020; 34: 101615.
- Cao B, Wang Y, Wen D, et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Eng J Med 2020; 382: 1787-1799.
- Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discover Therap2020; 14: 58-60.
- Foolad F, Aitken SL, Shigle TL, et al. Oral versus aerosolized ribavirin for the treatment of respiratory syncytial virus infections in hematopoietic cell transplant recipients. Clin Infect Dis 2019; 68: 1641-1649.
- Rosa SGV, Santos WC. Clinical trials on drug repositioning for COVID-19 treatment. Revista Panamericana de Salud Pública 2020; 44: e40.
- Xu X, Han M, Li T, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proceedings of the National Academy of Sciences2020; 117: 10970-10975.
- Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science2020; 368: 630-633.
- Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B. 2020.
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends2020; 14: 69-71.
- Yang Y, Islam MS, Wang J, et al. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci2020; 16: 1708.
- Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci2020; 134: 543-545.
